Description
ExcelTaq™ Taq DNA Polymerase is a recombinant thermo-stable Taq DNA polymerase expressed and purified from an E. coli strain carrying the cloned gene. With a high DNA synthesis rate and high thermo-stability, ExcelTaq™ Taq DNA Polymerase is suitable for common and specialized PCR applications.
Features
5'→3' DNA polymerase activity
5'→3' exonuclease activity
No detectable 3'→5' exonuclease (proofreading) activity
Generates PCR products with 3’-dA overhangs
Thermo-stable – half-life lasts for more than 40 min at 95°C
Applications
Routine PCR
Amplification of DNA fragments up to 8 kb
Generation of PCR products for TA cloning
DNA labeling
Storage
-20°C for 24 months
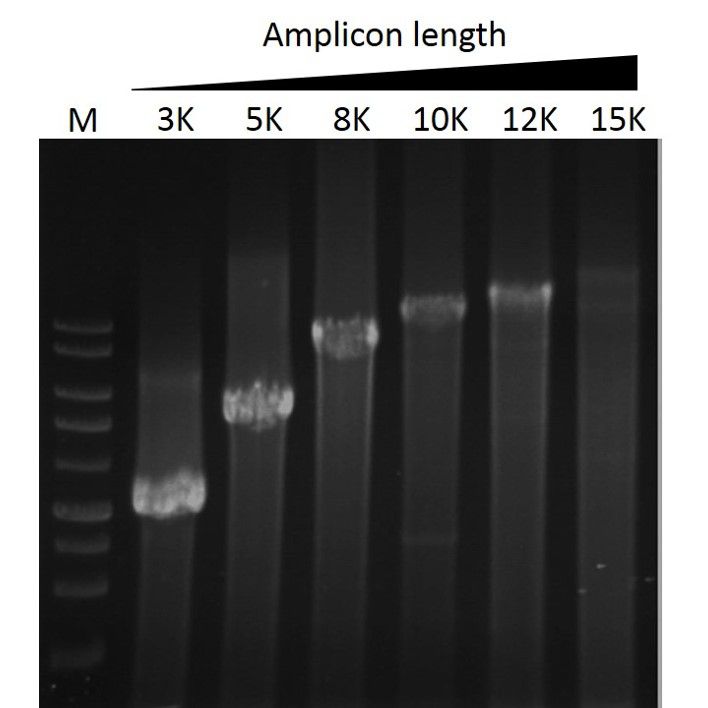
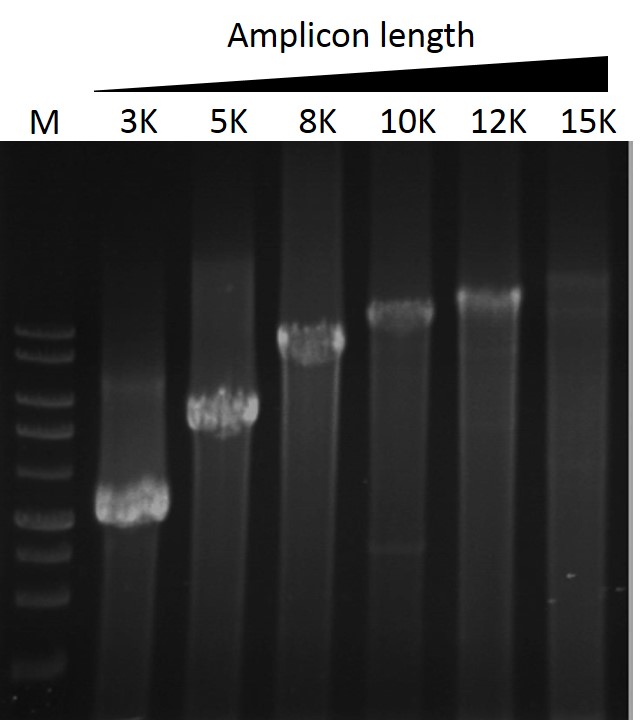
Elongation capability
ExcelTaq™ Taq DNA Polymerase can amplify PCR products from λDNA up to 15 kb (M: DM3100).
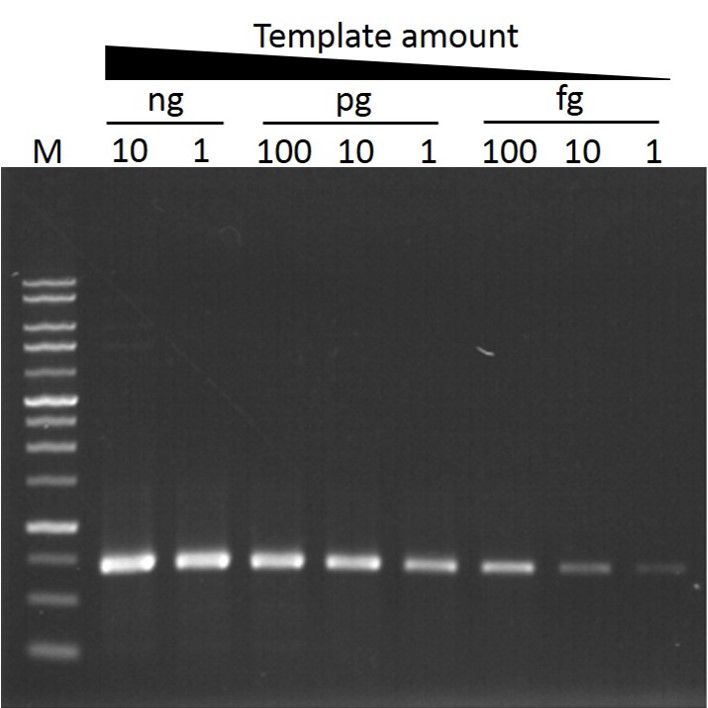
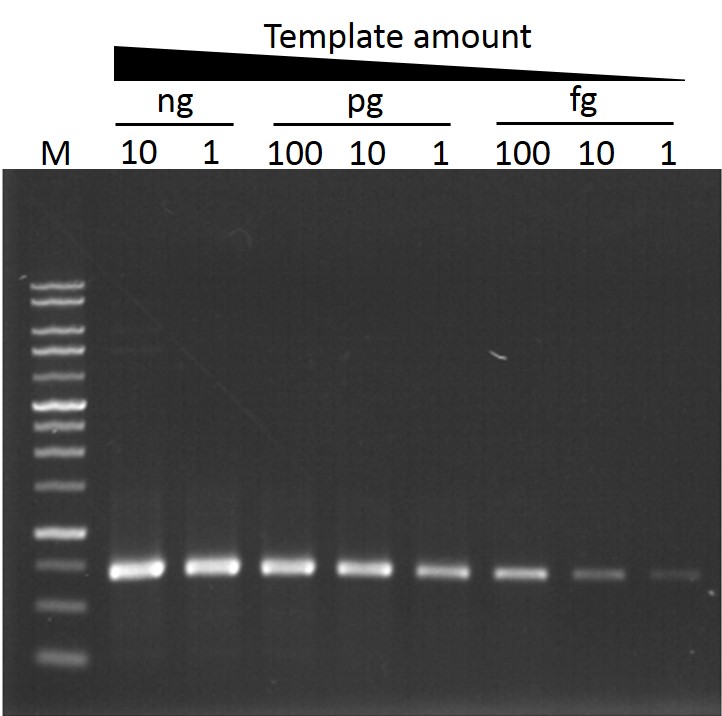
Sensitivity
ExcelTaq™ Taq DNA Polymerase can amplify PCR products from as little as 1 pg of template DNA (M: DM3100).
Contents
|
Storage Buffer
20 mM Tris-HCl (pH 8.0), 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, stabilizer, 50% (v/v) glycerol
10X Taq buffer
200 mM Tris-HCl (pH 8.8 at 25°C), 100 mM KCl, 100 mM (NH4)2SO4, 20 mM MgCl2, 1% Triton X-100
Unit Definition
One unit is defined as the amount of enzyme that will incorporate 10 nmol of dNTP into acid-insoluble material in 30 minutes at 74°C.
Storage
-20°C for 24 months
Recommended PCR Condition
Template | 1 – 150 ng |
Forward primer | 0.1 – 0.5 µM |
Reverse primer | 0.1 – 0.5 µM |
10X Taq Buffer | 5 µl |
dNTPs | 0.2 mM (each) |
Taq DNA Polymerase | 0.25 µl (1.25U) |
ddH2O | to 50 µl |
Total volume | 50 µl |
Recommended PCR Program
Steps | Temp. | Time | Cycles |
Template denature | 94°C | 2 min | 1 |
Denature | 94°C | 30 sec | 25-40 |
Annealing | 50-68°C* | 30 sec | |
Extension | 72°C | 30 sec/kb | |
Final extension | 72°C | 1 min | 1 |
*Optimal PCR condition varies according to primers’ thermodynamic properties.
The inhibitory effects of PGG and EGCG against the SARS-CoV-2 3C-like protease
PMCID: PMC7787066
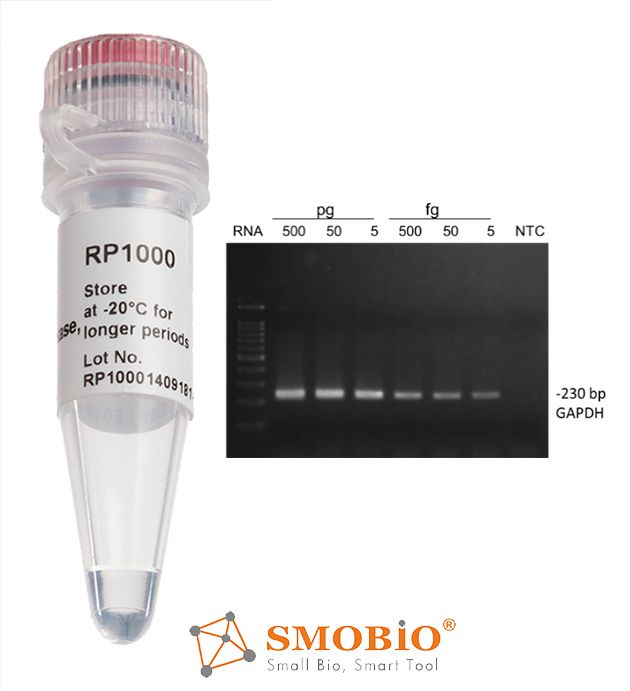
[RP1000] ExcelRT™ Reverse Transcriptase
High yield
Thermostable, up to 50°C, during first strand synthesis
High processivity, generating cDNA up to 8 kb
Reduced RNase H ribonuclease activity
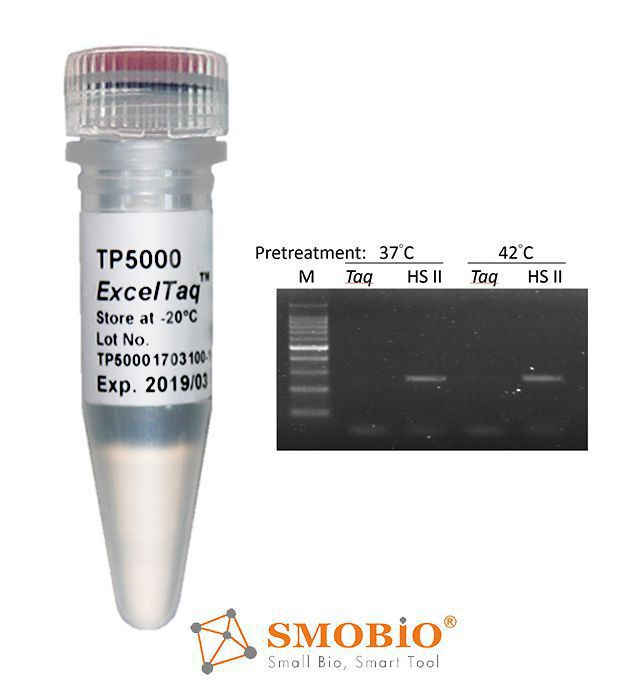
[TP5000] ExcelTaq™ Hot Start II DNA Polymerase
Aptamer-based hot start PCR
Reversible enzyme inactivation
Omits extra enzyme activation step
Convenient for room temperature PCR set-up
High yield and specificity of target amplicons
Wide range of amplicon length (up to 10 kb)
High sensitivity (as low as 1 fg of plasmid)
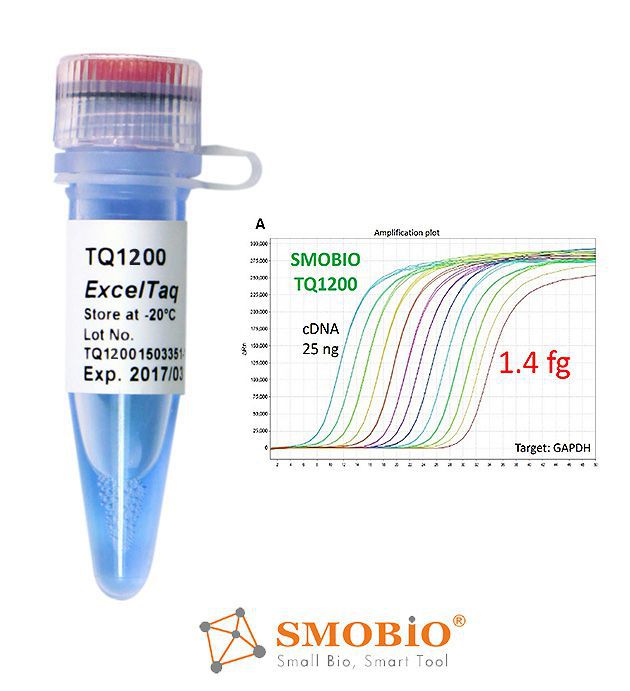
[TQ1200] ExcelTaq™ 2X Fast Q-PCR Master Mix (SYBR, no ROX)
High Stability
Fast Hot Start
High Sensitivity
Low Background / High Specificity
Suitable for Fast Program
Smart Blue Contrast Dye
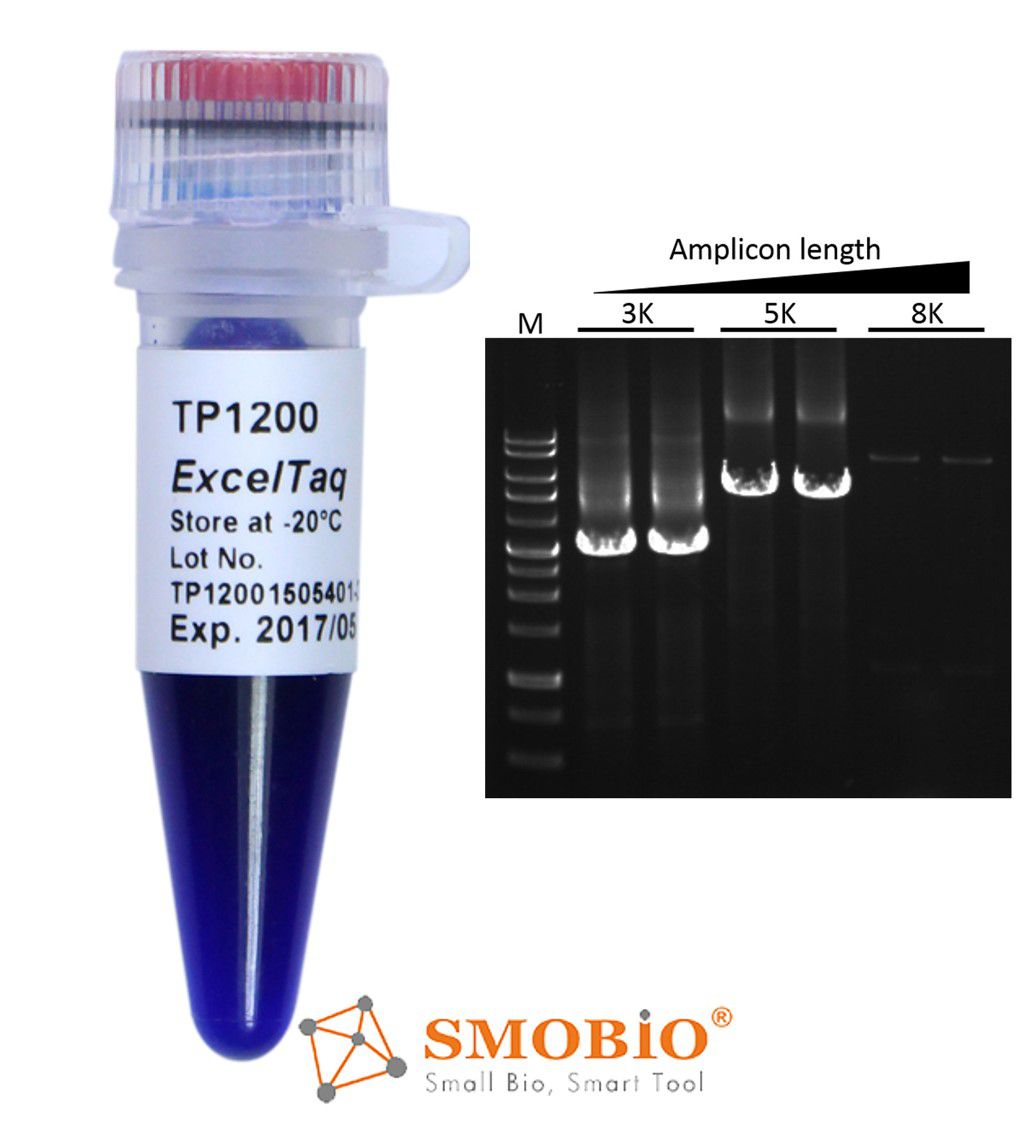
[TP1200] ExcelTaq™ 5X PCR Master Dye Mix
5’→3’ DNA polymerase activity
No detectable 3'→5' exonuclease (proofreading) activity
Generates PCR products with 3'-dA overhangs
High yield PCR
High reproducibility
Reduced pipetting errors
Includes tracking dye for direct loading after PCR

![[TP1000] ExcelTaq™ Taq DNA Polymerase, 5 U/μl, 500 U](/web/image/product.template/316/image?unique=6676958)
![[TP1000] ExcelTaq™ Taq DNA Polymerase, 5 U/μl, 500 U](/website/image/product.template/316/image/90x90)