Description
The G-HiFi™ DNA Polymerase is a new genetically modified, recombinant DNA polymerase suitable for GC-rich templates that are difficult to amplify. The fidelity of G-HiFi™ DNA Polymerase is 70 times higher than that of Taq DNA polymerase. The high extension rate of G-HiFi™ DNA Polymerase is achieved by blending the DNA polymerase with an elongation enhancer. The optimized 5X G-HiFi™ Buffer includes special ingredients that suppress non-specific amplification as well as plateau effect produced by conventional PCR. With the optimized 5X G-HiFi™ Buffer, G-HiFi™ DNA Polymerase is capable to amplify most templates, such as longer targets (up to 40 kb from lambda DNA) and that contain GC-rich sequences.
Features
5’→3’ DNA polymerase activity
3’→5’ exonuclease (proofreading) activity
Suitable for GC-rich templates
High reaction rate: 7 seconds/kb
High fidelity: 70 times higher than Taq polymerase
Generates blunt end amplicons
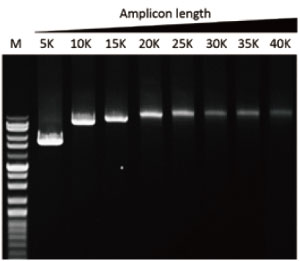
Vast elongation capability (up to 40 kb)
Thermo-stable for more than 10 hrs at 95°C
Storage
[TF3000] G-HiFi™ DNA Polymerase
-20°C for 24 months
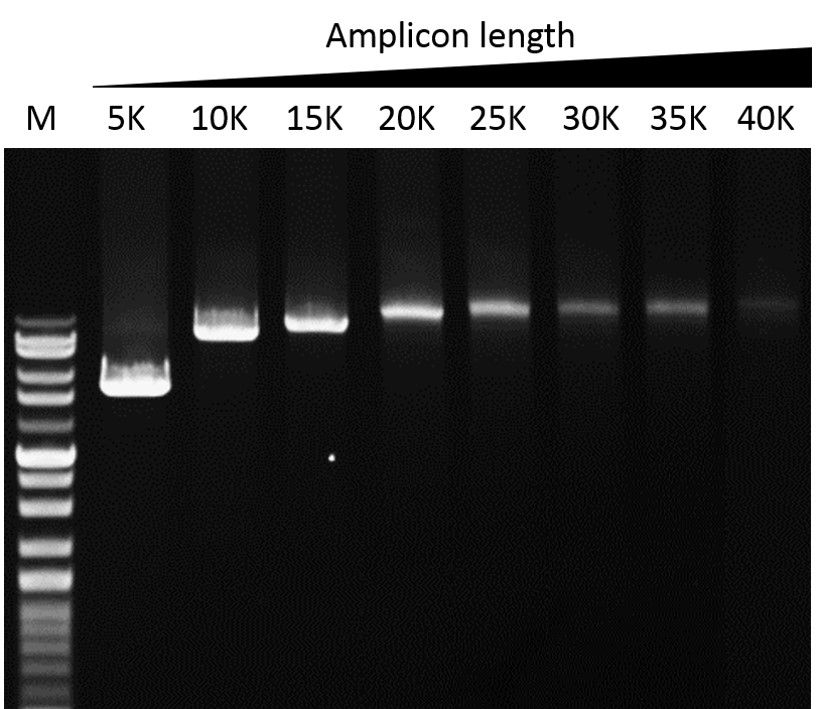
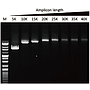
Elongation capability
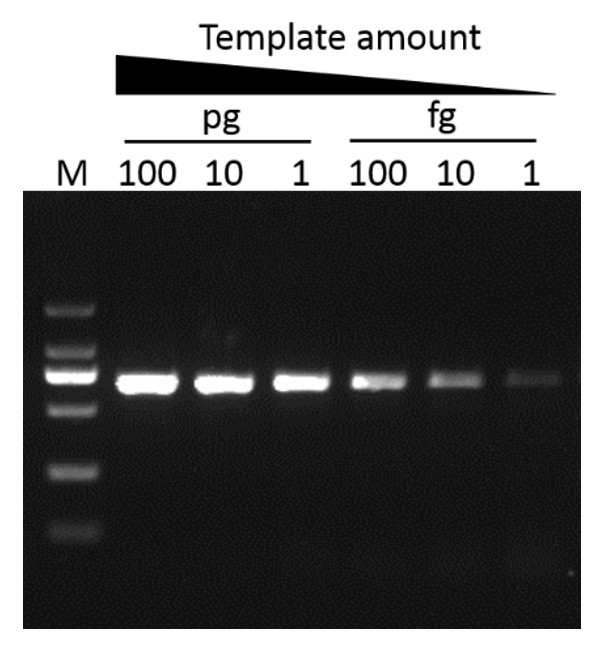
G-HiFi™ DNA Polymerase’s high processability enables reliable amplification of λDNA up to 40 kb in length (M: DM5100).
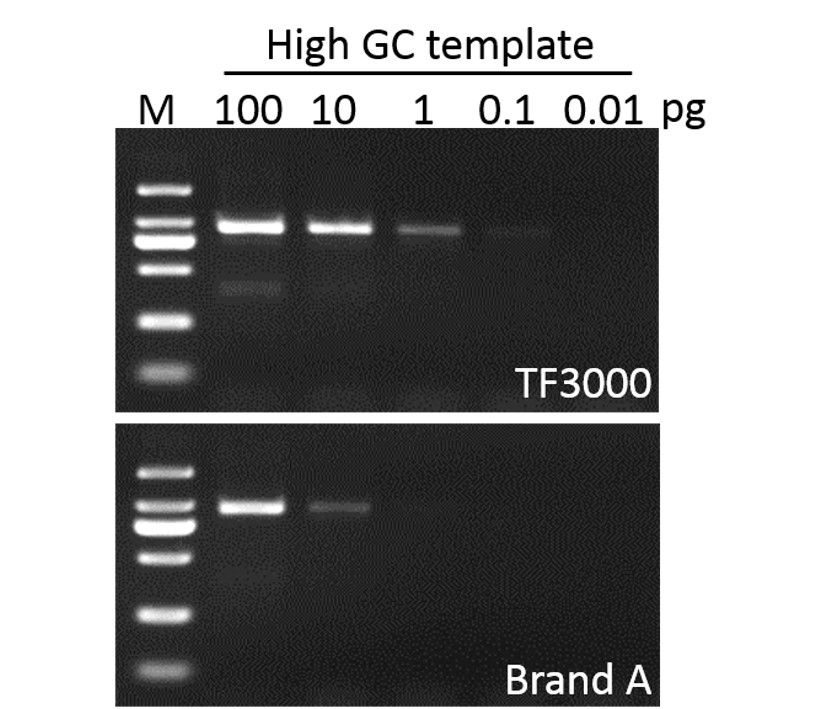
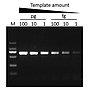
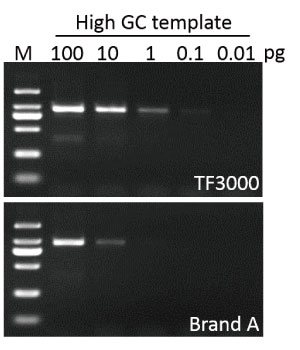
Sensitivity
G-HiFi™ DNA Polymerase performs higher sensitivity for high GC content templates (GC: 71%) compare to high fidelity DNA Polymerase from Brand A (M: DM2000).
[TF3000] G-HiFi™ DNA Polymerase
Contents
|
Storage Buffer
50 mM Tris-HCl (pH 8.0), 50 mM KCl, 0.1 mM EDTA, 1 mM DTT, stabilizer, 50% (v/v) glycerol
Unit Definition
One unit is defined as the amount of enzyme that will incorporate 10 nmol of dNTP into acid-insoluble material in 30 minutes at 74°C.
Storage
-20°C for 24 months
What are the differences between SMO-HiFi™ (TF1000) and G-HiFi™ (TF3000)?
Both SMO-HiFi™ (TF1000) and G-HiFi™ (TF3000) are new genetically modified, recombinant DNA polymerase with fidelity 70 times higher than Taq DNA polymerase during amplification.
The accelerated extension rate of G-HiFi™ DNA Polymerase (TF3000) is attained through a unique combination with an elongation enhancer.
Furthermore, TF3000, equipped with an optimized 5x G-HiFi buffer, exhibits the ability to efficiently synthesize long target (up to 40 kb from lambda DNA) and GC-rich DNA templates that are typically challenging to amplify.
Recommended PCR Condition
[TF3000] G-HiFi™ DNA Polymerase
Template | 1 – 150 ng |
Forward primer | 0.1 – 0.5 µM* |
Reverse primer | 0.1 – 0.5 µM* |
5X G-HiFi™ Buffer | 10 µl |
dNTPs Mix (2 mM each) | 5 µl |
G-HiFi™ DNA Polymerase | 0.5 – 1 unit** |
H2O | to 50 µl |
Total volume | 50 µl |
*When amplifying products ≧ 10 kb in length, use primers at a final concentration of 0.1 μM each.
** When amplifying products ≦ 2 kb in length, use 0.5 unit of G-HiFi™ DNA Polymerase.
Recommended PCR Program
For ≦ 10 kb products
Select primers with a Tm value of ≧55°C. 20- to 25-mer primers are suitable, or those greater than 25-mer in length may provide optimal results.
Steps | Temp. | Time | Cycles |
Template denature | 98°C | 2 min | 1 |
Denature | 98°C | 10 sec | 25-40 |
Annealing | 50-68°C | 15 sec | |
Extension | 68°C | 10-30 sec/kb | |
Final extension | 68°C | 1 min | 1 |
*Optimal PCR condition varies according to primers’ thermodynamic properties.
For ≧ 10 kb products
Select primers with a Tm value of ≧ 65°C. 25- to 35-mer primers are suitable. Avoid high GC-content at the 3' end of each primer.
Steps | Temp. | Time | Cycles |
Denature | 98°C | 10 sec | 25-40 |
Extension | 68°C | 10-30 sec/kb |
For GC-rich templates:
|
Steps |
Temp. |
Time |
Cycles |
|
Template denature |
98°C |
2 min |
1 |
|
Denature |
98°C |
10 sec |
25-40 |
|
Extension |
68°C |
10-30 sec/kb |
Samaneh Ghanbari, Elham Bayat, Masoumeh Azizi, Pezhman Fard-Esfahani, Mohammad Hossein Modarressi, and Fatemeh Davami
Appl Microbiol Biotechnol. 2022 Dec 20 : 1–15. doi: 10.1007/s00253-022-12322-1 [Epub ahead of print]
PMCID: PMC9763083
AFEAP cloning: a precise and efficient method for large DNA sequence assembly
Fanli Zeng,1 Jinping Zang,1 Suhua Zhang,2 Zhimin Hao,1 Jingao Dong,corresponding author1 and Yibin Lincorresponding author3
BMC Biotechnol. 2017; 17: 81. Published online 2017 Nov 14. doi: 10.1186/s12896-017-0394-x
PMCID: PMC5686892

Gel electrophoresis
Staining amplicons with safe fluorescent dyes, following by observation under blue-light illuminator to minimize damage of DNA amplicons and maximize successful cloning efficiency.
Safe fluorescent dyes
[NS1000] FluoroVue™ Nucleic Acid Gel Stain (10,000X), 500 μl
[DS1000] FluoroStain™ DNA Fluorescent Staining Dye (Green, 10,000X), 500 μl
[DL5000] FluoroDye™ DNA Fluorescent Loading Dye (Green, 6X), 1 ml
Blue-light illuminator
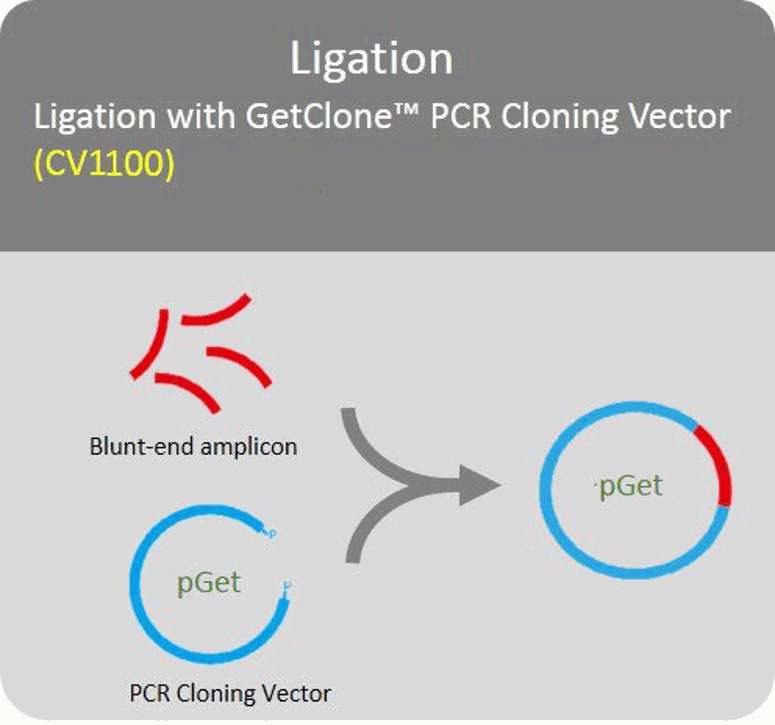
Ligation
Blund-end PCR amplicons can directly ligate with PCR cloning vector.
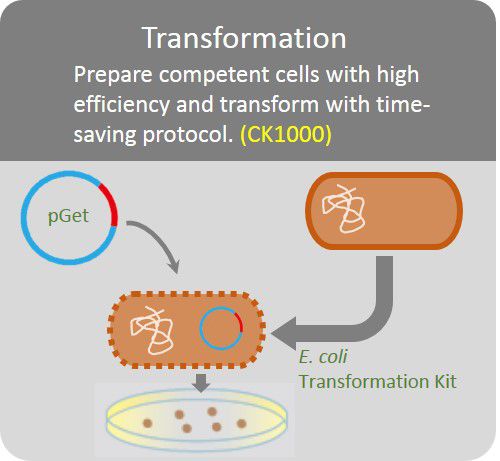
Transformation
Prepare competent cells with high efficiency and transform with time-saving protocol.
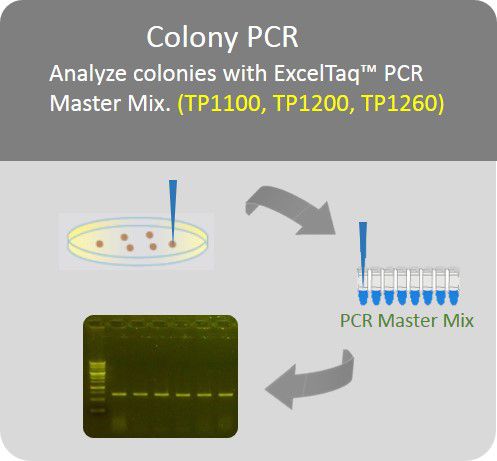
Colony PCR
Analyze colonies with PCR master mix to save preparation time.

![[TF3000] G-HiFi™ DNA Polymerase, 1 U/μl, 100 U](/web/image/product.template/387/image?unique=9bc6dac)
![[TF3000] G-HiFi™ DNA Polymerase, 1 U/μl, 100 U](/website/image/product.template/387/image/90x90)