Description
ExcelRT™ Reverse Transcription Kit is a complete, efficient and convenient kit to synthesize high quality first strand cDNA. This kit contains ExcelRT™ Reverse Transcriptase, which is able to synthesize the first strand cDNA at 37~50°C. The ExcelRT™ Reverse Transcriptase is a recombinant Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase, which is designed to reduce RNase H activity and create better thermal stability. This kit also contains RNAok™ RNase Inhibitor, which is active against RNase A, RNase B, and RNase C. This product is supplied with oligo (dT)20 and random hexamers, which are used to synthesize cDNA from poly(A) tailed mRNA and total RNA, respectively.
Features
Contains all components for reverse transcription
High yield
Thermostable, up to 50°C, during first strand synthesis
High processivity, generating cDNA up to 8 kb
Reduced RNase H ribonuclease activity
Application
Generation of first strand cDNA from total RNA or mRNA.
Suitable for generating cDNA from RNA with strong secondary structure which can be reduced at temperature up to 50°C.
Storage
-20°C for 24 months
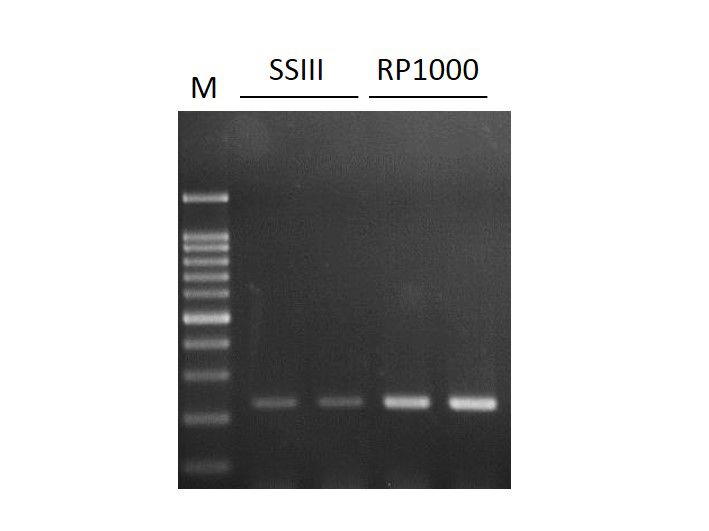
High yield
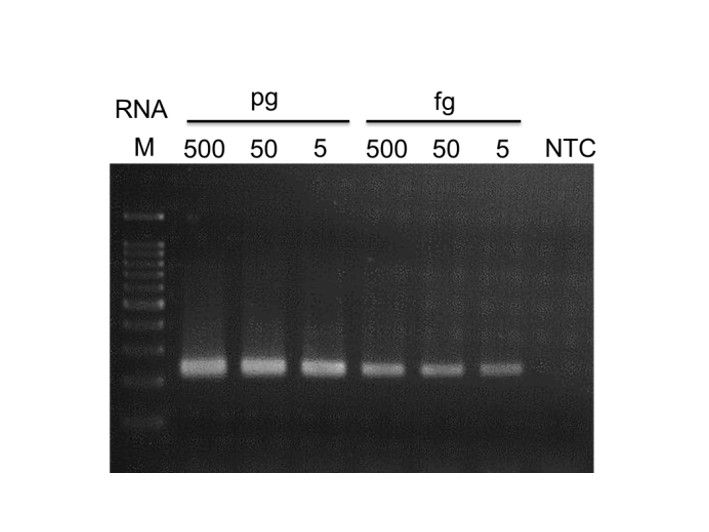
High sensitivity
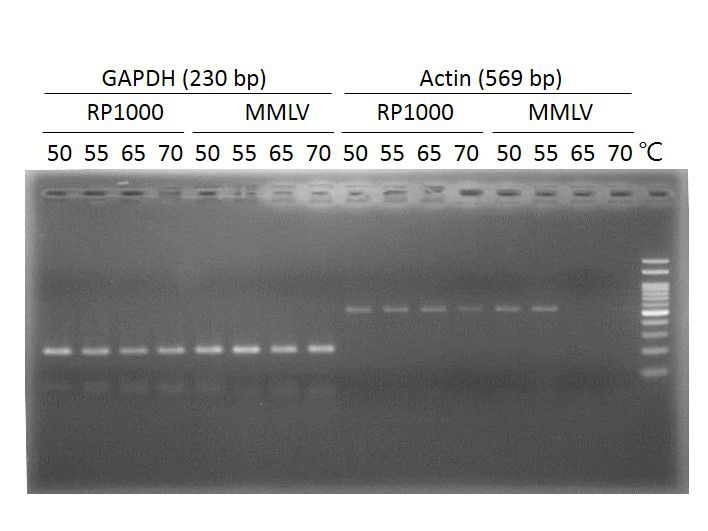
Thermostable, up to 50°C, during first strand synthesis
Contents
|
Storage Buffer
Reverse Transcriptase: 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 0.1 mM EDTA, 1 mM DTT, stabilizer and 50% (v/v) glycerol
RNase Inhibitor: 40 mM HEPES-KOH (pH 7.5), 100 mM KCl, 8 mM DTT, 0.1 mM EDTA, stabilizer and 50% (v/v) glycerol
5X RT buffer (DTT)
250 mM Tris-HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2 and 50 mM DTT
Storage
-20°C for 24 months
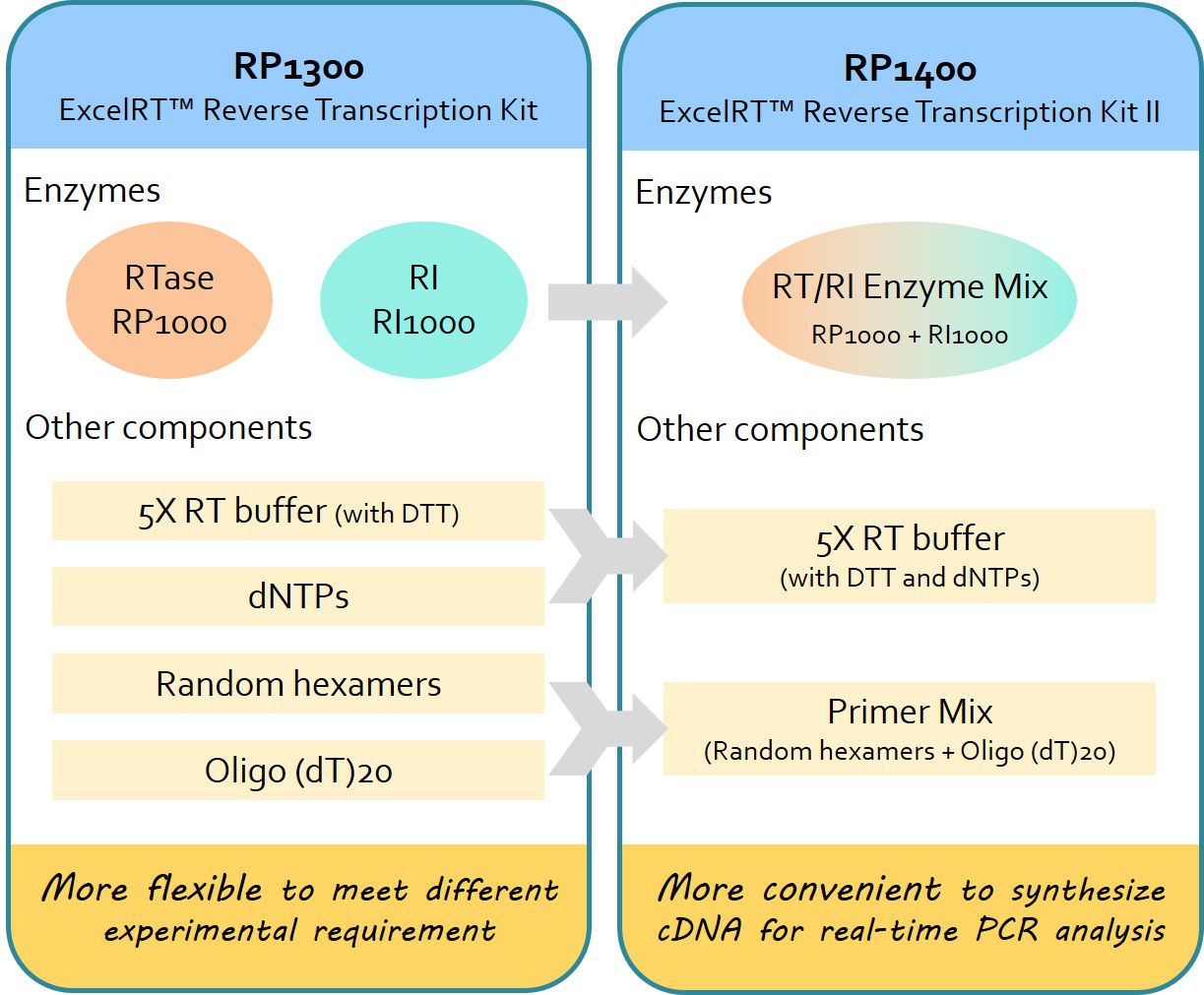
What is the difference between ExcelRT™ Reverse Transcription Kit (RP1300) and ExcelRT™ Reverse Transcription Kit II (RP1400)?
ExcelRT™ Reverse Transcription Kits contain all components to synthesize high quality first strand cDNA. The components in ExcelRT™ Reverse Transcription Kit (RP1300) and ExcelRT™ Reverse Transcription Kit II (RP1400) are the same.
The components in RP1300 are separated. This make RP1300 more flexible to fit different experimental requirements.
Selected components in RP1400 are pre-mixed. This make RP1400 more convenient to use.
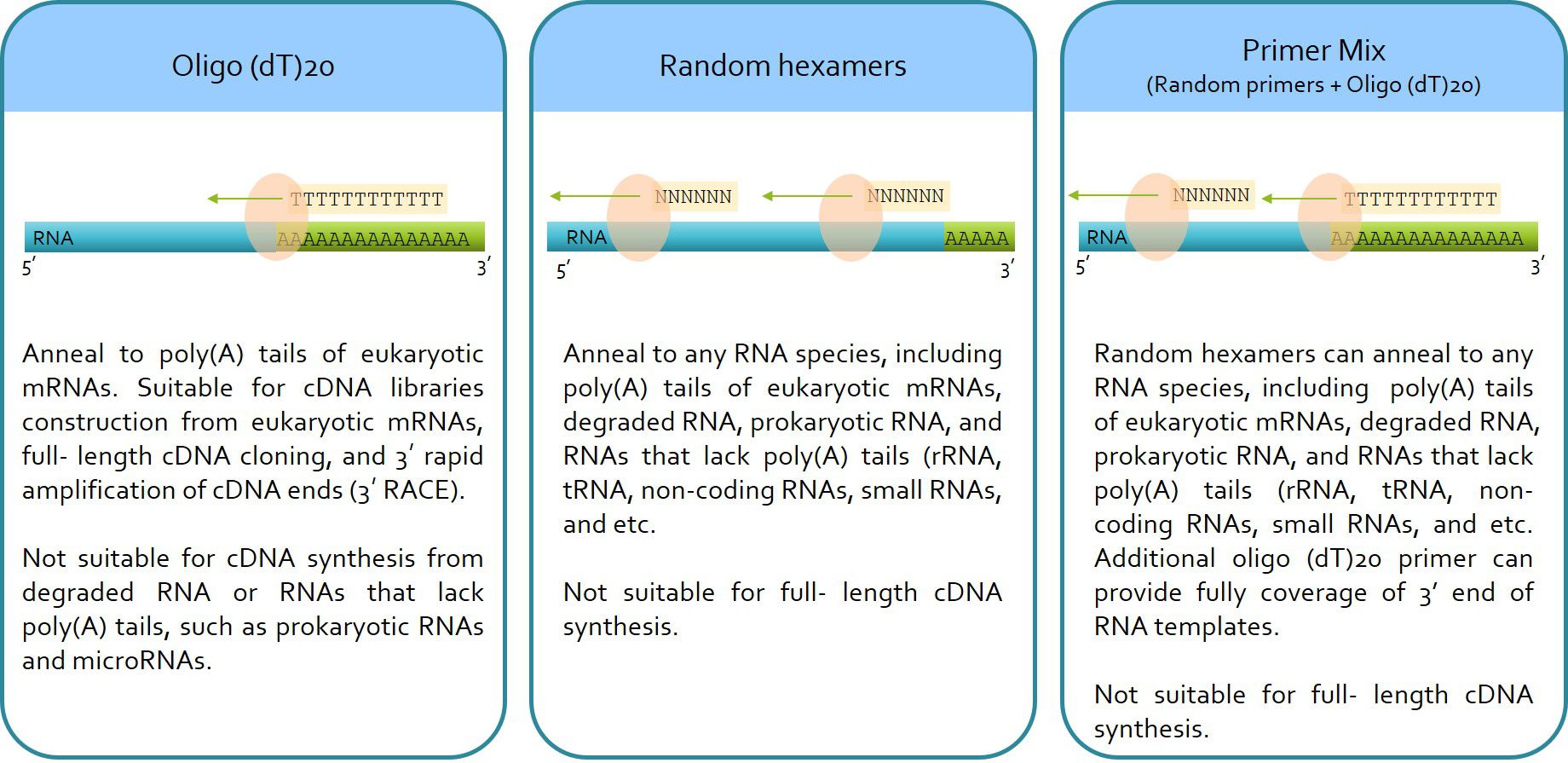
ExcelRT™ Reverse Transcription Kit (RP1300) contains Random hexamers and Oligo(dT)20, how do I choose?
You can choose oligo d(T)20, Random hexamers, or both depending on the RNA template and the downstream applications.
Oligo (dT)20 anneals to poly(A) tails of eukaryotic mRNAs. It is suitable for cDNA libraries construction from eukaryotic mRNAs, full- length cDNA cloning, and 3′ rapid amplification of cDNA ends (3′ RACE). It is not suitable for cDNA synthesis from degraded RNA or RNAs that lack poly(A) tails, such as prokaryotic RNAs and microRNAs.
Narges Karami, Adeleh Taei, Seyedeh Nafiseh Hassani, Nazanin Alizadeh, Poopak Eftekhari-Yazdi, Fatemeh Hassani
Sci Rep. 2025 Feb 11;15(1):5006. doi: 10.1038/s41598-025-89460-9.
PMCID: PMC11811121
The ratio of COL2A1:COL1A1 in dartos tissue patients with hypospadias
Prahara Yuri, Hotman Christinus, Zico Yusuf Alfarizi, Medina Ndoye
BMC Urol. 2025 Jan 4;25(1):2. doi: 10.1186/s12894-024-01688-1.
PMCID: PMC11699663
Importance of LINC00852/miR-145-5p in breast cancer: a bioinformatics and experimental study
Abbas Shakoori, Asghar Hosseinzadeh, Nahid Nafisi, Ramesh Omranipour, Leyla Sahebi, Nazanin Hosseinkhan, Mohsen Ahmadi, Soudeh Ghafouri-Fard, Maryam Abtin
Discov Oncol. 2024 Nov 18;15(1):672. doi: 10.1007/s12672-024-01553-5.
PMCID: PMC11574217
Dousti M, Mousavi Jafaripour E, Ahmadzadeh M, Falahi Charkhabi N, Ahmadzadeh M.
Sci Rep. 2024 Dec 28;14(1):31011. doi: 10.1038/s41598-024-82167-3.
PMCID: PMC11680889
Zafarani F, Soleimani Mehranjani M, Ghaffari F, Shahhoseini M, Shariatzadeh SMA.
Int J Fertil Steril. 2024 Oct 30;18(4):344-351. doi: 10.22074/ijfs.2024.2026081.1655.
PMCID: PMC11589967
Rattanapan Y, Nongwa K, Supanpong C, Satsadeedat C, Sai-Ong T, Kooltheat N, Chareonsirisuthigul T.
J Clin Med Res. 2024 Dec;16(11):536-546. doi: 10.14740/jocmr6099. Epub 2024 Nov 30.
PMCID: PMC11614410
Fatemeh Mardani-Korrani, Rayhaneh Amooaghaie, Alimohammad Ahadi, Mustafa Ghanadian
BMC Plant Biol. 2024 Aug 24;24(1):798. doi: 10.1186/s12870-024-05502-w.
PMCID: PMC11344448
Nooshafarin Shirani, Roohallah Mahdi-Esferizi, Reza Eshraghi Samani, Shahram Tahmasebian, Hajar Yaghoobi
Cancer Rep (Hoboken). 2024 Jun;7(6):e2114. doi: 10.1002/cnr2.2114.
PMCID: PMC11182701
Sajad Jeddi, Nasibeh Yousefzadeh, Maryam Zarkesh, Khosrow Kashfi, Asghar Ghasemi
Front Pharmacol. 2024 Mar 27:15:1369379. doi: 10.3389/fphar.2024.1369379. eCollection 2024.
PMCID: PMC11004245
Ali Akbar Soleimani, Nafiseh Shokri, Mohammad Elahimanesh, Payam Mohammadi, Najmeh Parvaz, Masoomeh Bakhshandeh, Mohammad Najafi
Endocrinol Diabetes Metab. 2024 Jan; 7(1): e465. Published online 2023 Dec 15. doi: 10.1002/edm2.465
PMCID: PMC10782052
Banafsheh Rastegari, Atefe Ghamar Talepoor, Shahdad Khosropanah, Mehrnoosh Doroudchi
ACS Omega. 2024 Jan 9; 9(1): 658–674. Published online 2023 Dec 18. doi: 10.1021/acsomega.3c06389
PMCID: PMC10785661
Effect of drought stress on natural rubber biosynthesis and quality in Taraxacum kok-saghyz roots
Seyed Shahab Hedayat Mofidi, Mohammad Reza Naghavi, Manijeh Sabokdast, Parisa Jariani, Meisam Zargar, Katrina Cornish
PLoS One. 2024; 19(1): e0295694. Published online 2024 Jan 22. doi: 10.1371/journal.pone.0295694
PMCID: PMC10802950
Maryam Abtin, Nahid Nafisi, Asghar Hosseinzadeh, Sepideh Kadkhoda, Ramesh Omranipour, Leyla Sahebi, Masoumeh Razipour, Soudeh Ghafouri-Fard, Abbas Shakoori
Noncoding RNA Res. 2024 Jun; 9(2): 367–375. Published online 2024 Jan 26. doi: 10.1016/j.ncrna.2024.01.016
PMCID: PMC10950563
Assessment of expression of calcium signaling related lncRNAs in epilepsy
Mohammad Taheri, Ashkan Pourtavakoli, Solat Eslami, Soudeh Ghafouri-Fard, Arezou Sayad
Sci Rep. 2023; 13: 17993. Published online 2023 Oct 21. doi: 10.1038/s41598-023-45341-7
PMCID: PMC10590428
Younes Yassaghi, Sajad Jeddi, Nasibeh Yousefzadeh, Khosrow Kashfi, Asghar Ghasemi BMC Cardiovasc Disord. 2023 Aug 21;23(1):411. doi: 10.1186/s12872-023-03425-2.
PMCID: PMC10441752
Mohammad Houshyar, Ali Asghar Saki, Mohammad Yousef Alikhani, Michael Richard Bedford, Meysam Soleimani, Farideh Kamarehei
Poult Sci. 2023 Aug 9;102(11):103014. doi:10.1016/j.psj.2023.103014. Online ahead of print.
PMCID: PMC10494260
Hamed Rezaie, Reza Alipanah-Moghadam, Farhad Jeddi, Cain C T Clark, Vahideh Aghamohammadi, Ali Nemati
Sci Rep. 2023 Sep 12;13(1):15074. doi: 10.1038/s41598-023-42177-z.
PMCID: PMC10497591
Nasibeh Yousefzadeh, Sajad Jeddi, Maryam Zarkesh, Khosrow Kashfi , Asghar Ghasemi. Sci Rep. 2023 Mar 10;13(1):4013. doi: 10.1038/s41598-023-31240-4.
PMCID: PMC10006425
Mohammad Rafiee, Fatemeh Amiri, Mohammad Hossein Mohammadi, Abbas Hajifathali. BMC Cancer. 2023 Mar 3;23(1):202. doi: 10.1186/s12885-023-10665-0.
PMCID: PMC9983186
Salvatore Villani, Giulia Dematteis, Laura Tapella, Mara Gagliardi, Dmitry Lim, Marco Corazzari, Silvio Aprile and Erika Del Grosso
Pharmaceuticals (Basel). 2023 Feb; 16(2): 298. Published online 2023 Feb 14. doi: 10.3390/ph16020298
PMCID: PMC9960049
Abolfazl Naimabadi, Ahmad Ghasemi, Mahnaz Mohtashami, Jafar Saeidi, Mehdi Bakaeian, Aliakbar Haddad Mashadrizeh, Mohsen Azimi-Nezhad, Ali Akbar Mohammadia
Heliyon. 2022 Dec; 8(12): e12414. Published online 2022 Dec 19. doi: 10.1016/j.heliyon.2022.e12414
PMCID: PMC9803783
Gene expression profile of placentomes and clinical parameters in the cows with retained placenta
Mehdi Moradi, Mahdi Zhandi, Mohsen Sharafi, Arvand Akbari, Mohammad Jafari Atrabi, and Mehdi Totonchi
BMC Genomics. 2022; 23: 760. Published online 2022 Nov 21. doi: 10.1186/ s12864-022-08989-5
PMCID: PMC9677913
Assessment of Expression of Regulatory T Cell Differentiation Genes in Autism Spectrum Disorder
Mohammadarian Akbari, Reyhane Eghtedarian, Bashdar Mahmud Hussen, Solat Eslami, Mohammad Taheri, Seyedeh Morvarid Neishabouri, Soudeh Ghafouri-Fard, Front Mol Neurosci. 2022 Jul 4;15:939224. doi: 10.3389/fnmol.2022.939224. eCollection 2022.
PMCID: PMC9289514
Negar Sarmadi, Hossein Poustchi, Fatemeh Ali Yari, Amir Reza Radmard, Sara Karami, Abbas Pakdel, Parisa Shabani, Ali Khaleghian, PLoS One. 2022 Apr 12;17(4):e0266227. doi: 10.1371/journal.pone.0266227. eCollection 2022.
PMCID: PMC9004768
Expression analysis of IFNAR1 and TYK2 transcripts in COVID-19 patients
Mohammadarian Akbari, Mehdi Akhavan-Bahabadi, Navid Shafigh, Afshin Taheriazam, Bashdar Mahmud Hussen, Arezou Sayad, Mohadeseh Fathi, Mohammad Taheri, Soudeh Ghafouri-Fard, Mohammad Fathi, Cytokine. 2022 May;153:155849. doi: 10.1016/j.cyto.2022.155849. Epub 2022 Mar 4.
PMCID: PMC8894869
Ali Akbar Soleimani, Ghasem Ghasmpour, Asghar Mohammadi, Masoomeh Gholizadeh, Borhan Rahimi Abkenar, Mohammad Najafi, Endocrinol Diabetes Metab. 2022 May 28;e351. doi: 10.1002/edm2.351.
PMCID: PMC9258994
Direct RNA Nanopore Sequencing of SARS-CoV-2 Extracted from Critical Material from Swabs
Davide Vacca, Antonino Fiannaca, Fabio Tramuto, Valeria Cancila, Laura La Paglia, Walter Mazzucco, Alessandro Gulino, Massimo La Rosa, Carmelo Massimo Maida, Gaia Morello, Beatrice Belmonte, Alessandra Casuccio, Rosario Maugeri, Gerardo Iacopino, Carmela Rita Balistreri, Francesco Vitale, Claudio Tripodo, Alfonso Urso, Life (Basel). 2022 Jan 4;12(1):69. doi: 10.3390/life12010069.
PMCID: PMC8778588
Atefeh Babaei, Reza Asadpour, Kamran Mansouri, Adel Sabrivand, Siamak Kazemi-Darabadi, Food Science & Nutrition March 2022. doi:10.1002/fsn3.2762
PMCID: PMC9094497
Up-regulation of FOXN3-AS1 in invasive ductal carcinoma of breast cancer patients
Samira Molaei Ramshe 1, Hamid Ghaedi 1, Mir Davood Omrani 2, Lobat Geranpayeh 3, Behnam Alipour 4, Soudeh Ghafouri-Fard, Heliyon
. 2021 Oct 14;7(10):e08179. doi: 10.1016/j.heliyon.2021.e08179. eCollection 2021 Oct.
PMCID: PMC8526775
Bita Hassani, Mohammad Taheri, Yazdan Asgari, Ali Zekri, Ali Sattari, Soudeh Ghafouri-Fard, * and Farkhondeh Pouresmaeili , Front Oncol. 2021; 11: 671418. Published online 2021 May 19. doi: 10.3389/fonc.2021.671418
PMCID: PMC8171254
Pelin Kaya, Sang R. Lee, Young Ho Lee, Sun Woo Kwon, Hyun Yang, Hye Won Lee, and Eui-Ju Hong1, Nutrients. 2019 Feb; 11(2): 410. Published online 2019 Feb 15. doi: 10.3390/nu11020410
PMCID: PMC6412318
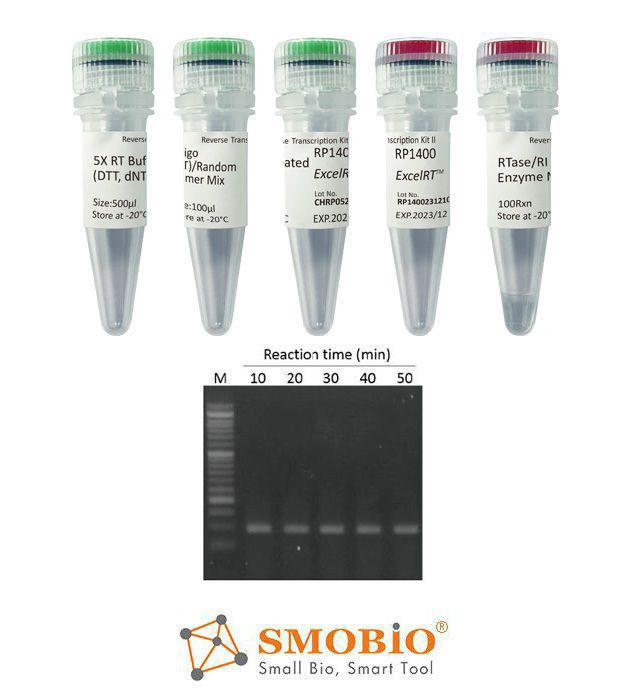
[RP1400] ExcelRT™ Reverse Transcription Kit II
Contains all components for reverse transcription
High yield
Thermostable, up to 50°C
Reduced RNase H ribonuclease activitySuitable for real-time PCR
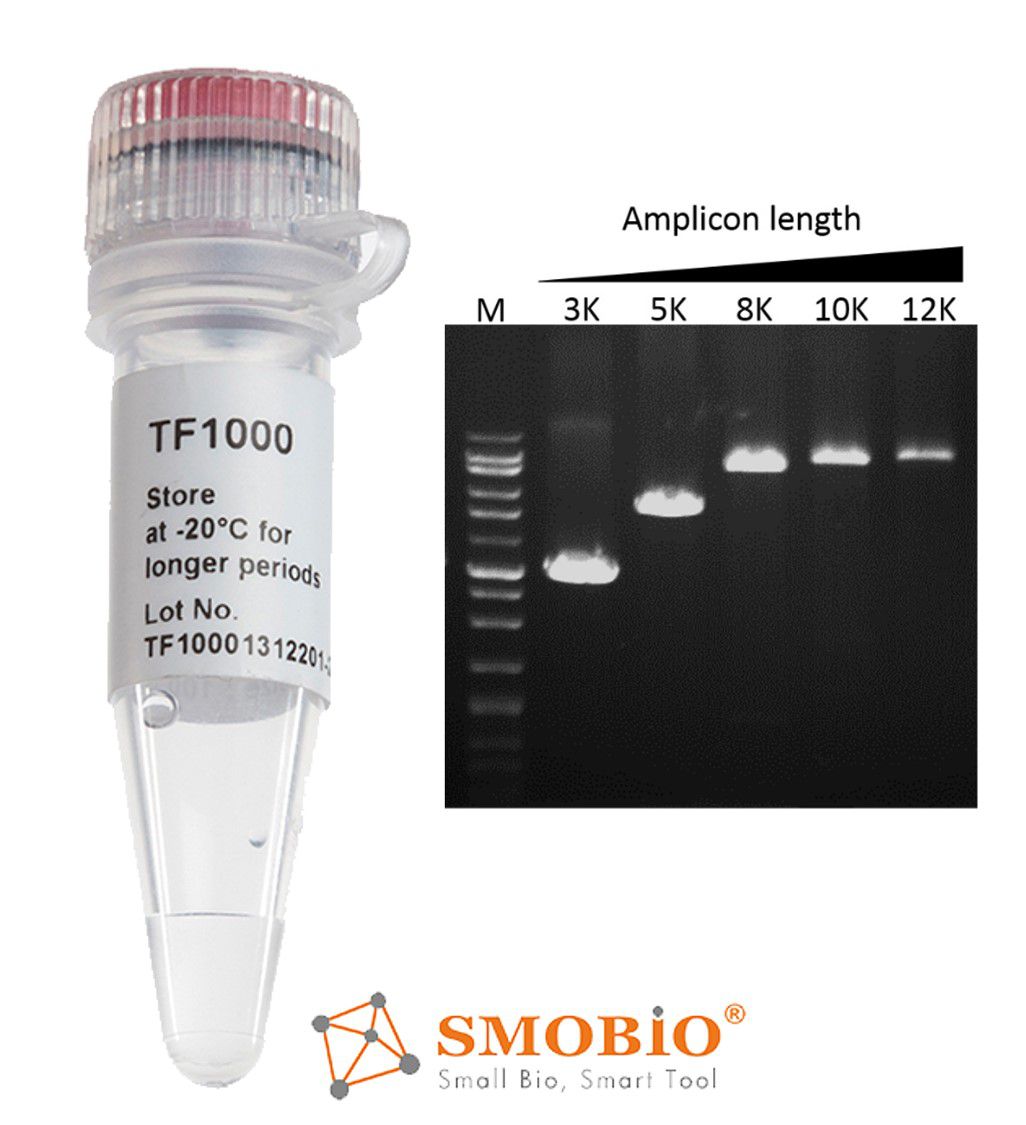
[TF1000] SMO-HiFi™ DNA Polymerase
5’→3’ DNA polymerase activity
3’→5’ exonuclease (proofreading) activity
High reaction rate—10 seconds/kb
High fidelity— 70 times higher than Taq polymerase
Generates blunt end amplicon
Thermo-stable for more than 10 hrs at 95°C.
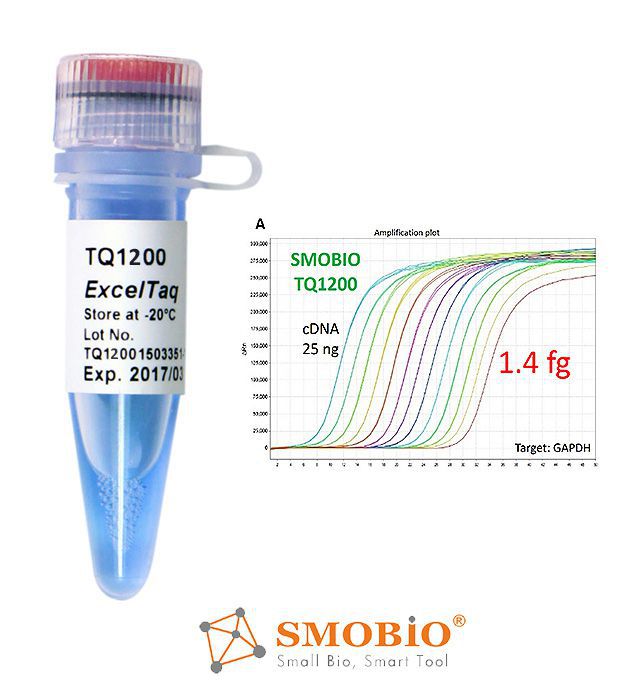
[TQ1200] ExcelTaq™ 2X Fast Q-PCR Master Mix (SYBR, no ROX)
High sensitivity and signal intensity
Compatible with fast PCR program
With smart blue contrast dye as a visual aid for reaction setup
Low background
High stability
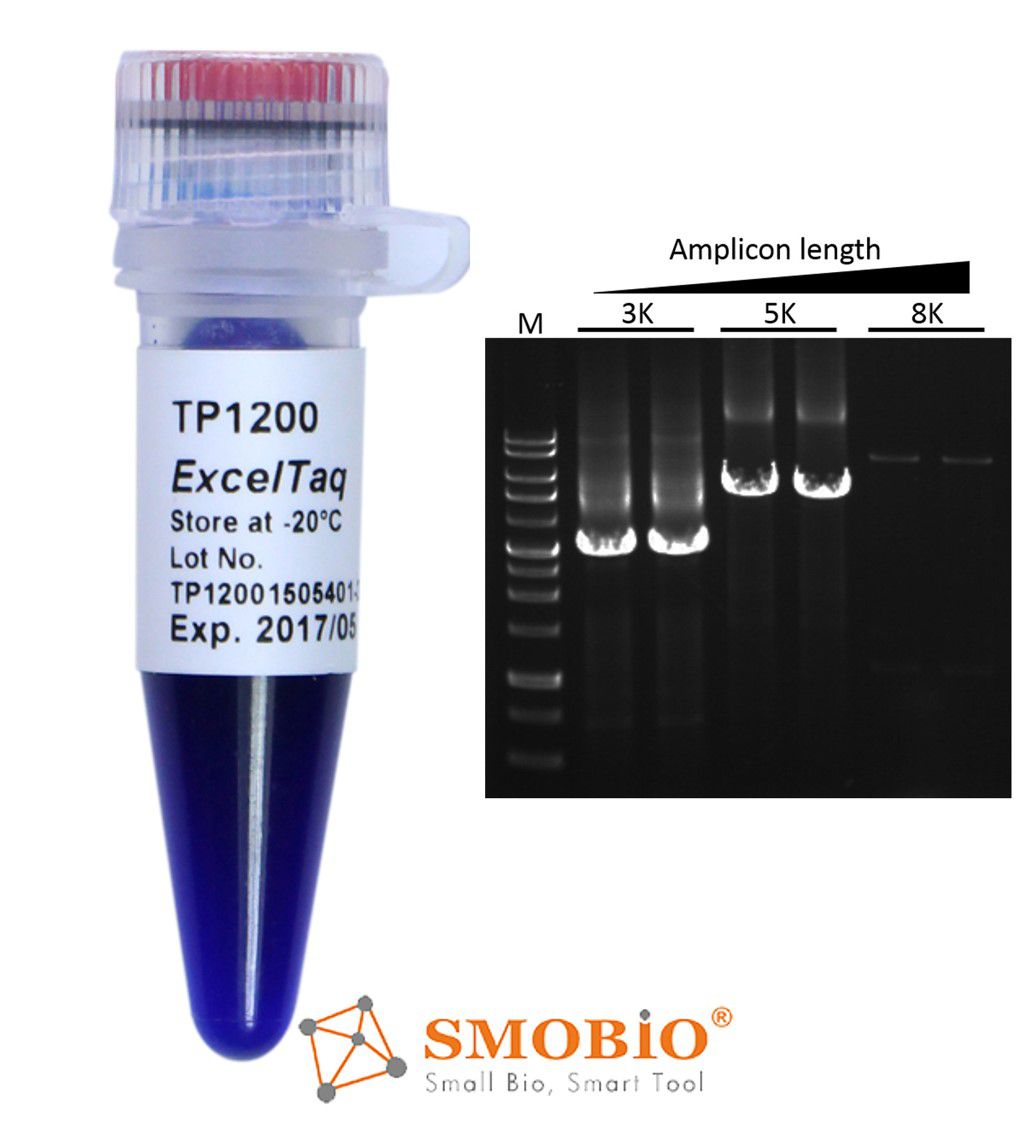
[TP1200] ExcelTaq™ 5X PCR Master Dye Mix
5’→3’ DNA polymerase activity
No detectable 3'→5' exonuclease (proofreading) activity
Generates PCR products with 3'-dA overhangs
High yield PCR
High reproducibility
Reduced pipetting errors
Includes tracking dye for direct loading after PCR

![[RP1300] ExcelRT™ Reverse Transcription Kit, 100 RXN](/web/image/product.template/273/image?unique=390b8cc)