Description
The ExcelRT™ Reverse Transcriptase is a recombinant Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase – an RNA dependent DNA polymerase capable of generating first strand cDNA using an RNA template. It is designed to reduce RNase H activity and create better thermal stability. The ExcelRT™ Reverse Transcriptase is able to routinely synthesize first strand cDNA >8 kb at 37~50°C.
Features
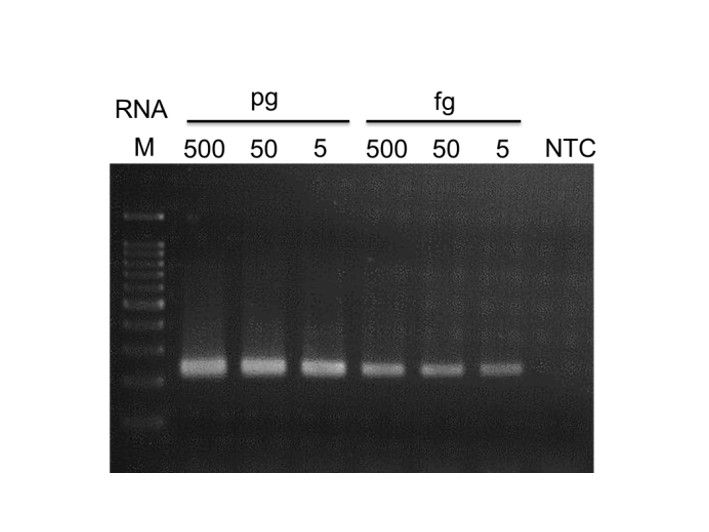
High yield
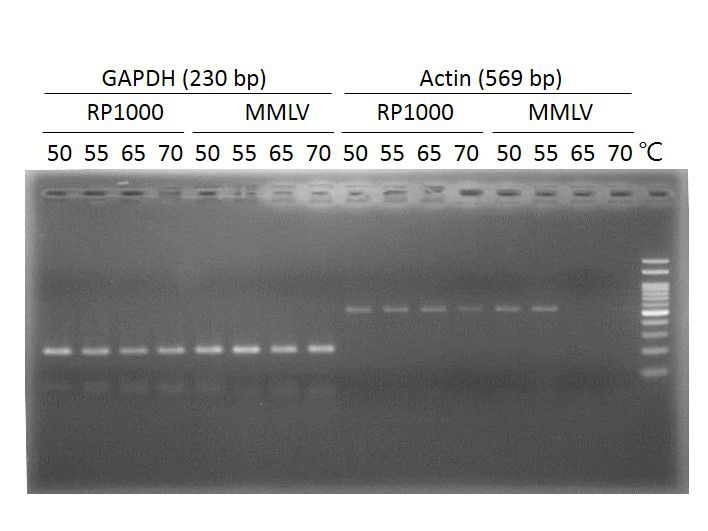
Thermostable, up to 50°C, during first strand synthesis
High processivity, generating cDNA up to 8 kb
Reduced RNase H ribonuclease activity
Application
Generation of first strand cDNA from total RNA or mRNA.
Suitable for generating cDNA from RNA with strong secondary structure which can be reduced at temperature up to 50°C.
Storage
-20°C for 24 months
High yield
Thermostable, up to 50°C, during first strand synthesis
High sensitivity
Contents
|
Storage Buffer
20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 0.1 mM EDTA, 1 mM DTT, stabilizer, 50% (v/v) glycerol
5X RT buffer
250 mM Tris-HCl (pH 8.3 at 25°C), 375 mM KCl and 15 mM MgCl2
Storage
-20°C for 24 months
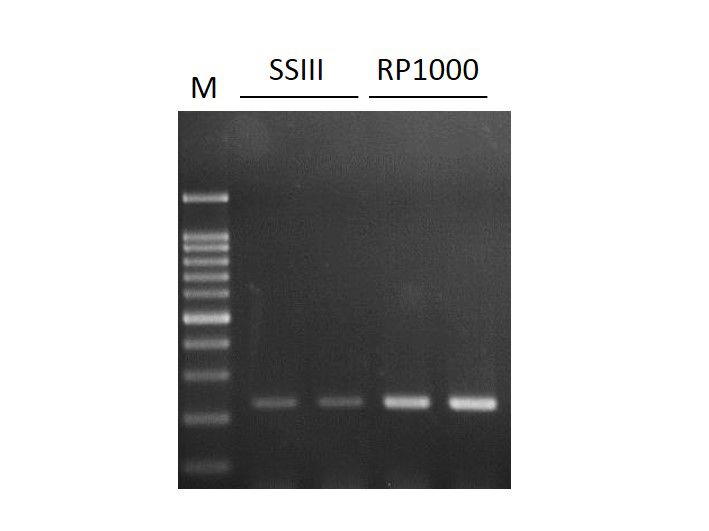
Can the reverse transcriptase RP1000 be diluted to one quarter for general use?
Probably, but it depends on the quantity and quality of RNA that individuals use. In most cases, the recommended amount of enzyme delivers the best result. Diluted usage may vary its effectiveness.
Bijorn Omar Balzamino, Lucia Dinice, Andrea Cacciamani, Agnese Re, Fabio Scarinci, Luca Bruno, Pamela Cosimi, and Alessandra Micera
Biomed Res Int. 2022; 2022: 7497816. Published online 2022 Dec 16. doi: 10.1155/2022/7497816
PMCID: PMC9788888
Romina Monzani, Mara Gagliardi, Nausicaa Clemente, Valentina Saverio, Elżbieta Pańczyszyn, Claudio Santoro, Nissan Yissachar, Annalisa Visciglia, Marco Pane, Angela Amoruso, and Marco Corazzari
Biology (Basel). 2022 Nov; 11(11): 1574. Published online 2022 Oct 27. doi: 10.3390/ biology11111574
PMCID: PMC9687175
Sulbactam-enhanced cytotoxicity of doxorubicin in breast cancer cells
Shao-hsuan Wen, Shey-chiang Su, Bo-huang Liou, Cheng-hao Lin, and Kuan-rong Lee, Cancer Cell Int. 2018; 18: 128. Published online 2018 Sep 4. doi: 10.1186/s12935-018-0625-9
PMCID: PMC6123926
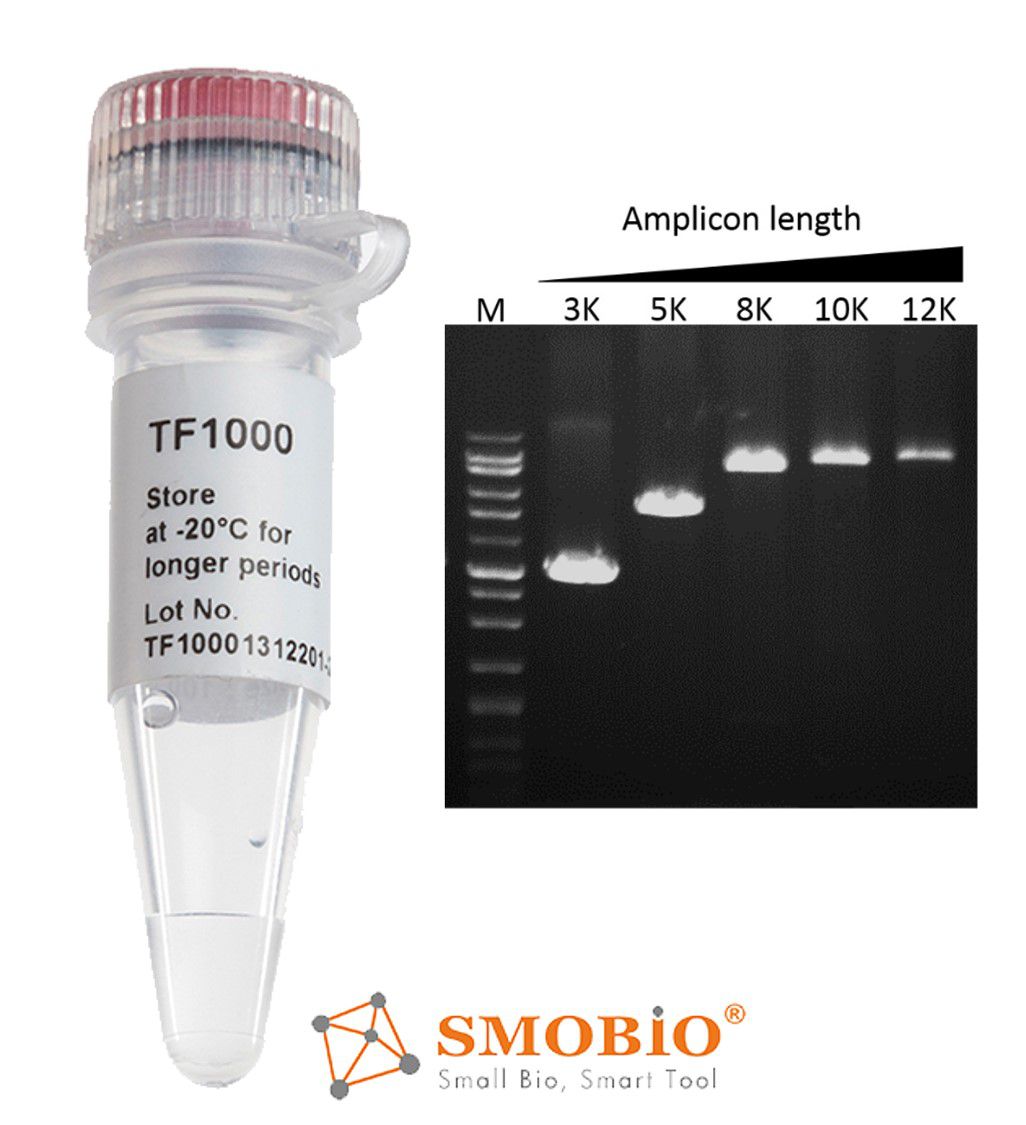
[TF1000] SMO-HiFi™ DNA Polymerase
5’→3’ DNA polymerase activity
3’→5’ exonuclease (proofreading) activity
High reaction rate—10 seconds/kb
High fidelity— 70 times higher than Taq polymerase
Generates blunt end amplicon
Thermo-stable for more than 10 hrs at 95°C.
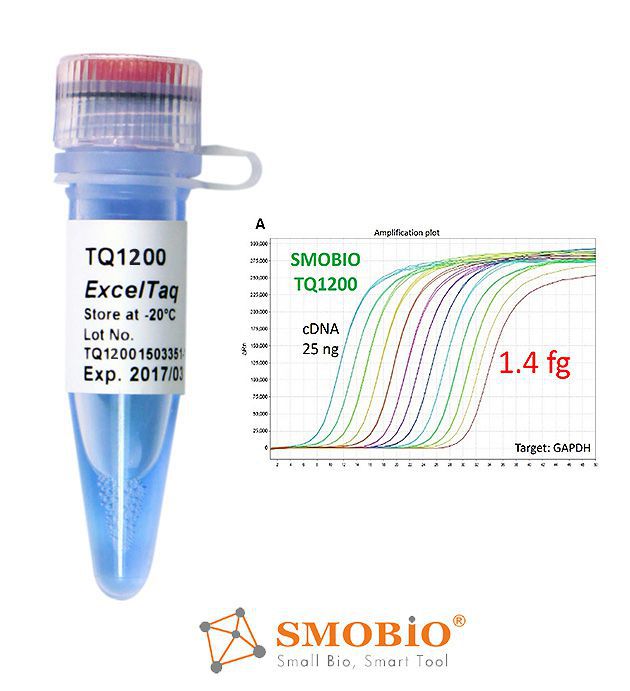
[TQ1200] ExcelTaq™ 2X Fast Q-PCR Master Mix (SYBR, no ROX)
High sensitivity and signal intensity
Compatible with fast PCR program
With smart blue contrast dye as a visual aid for reaction setup
Low background
High stability
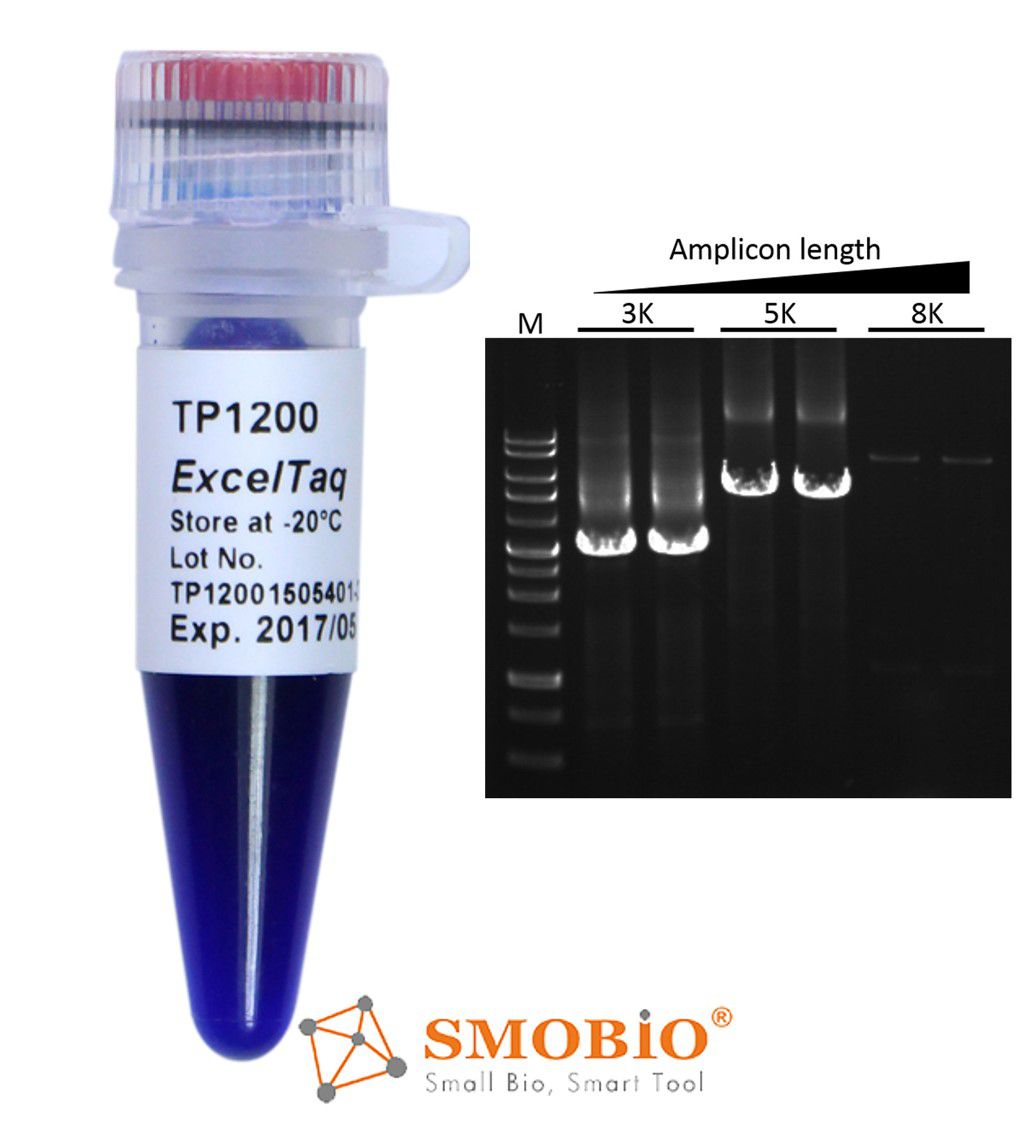
[TP1200] ExcelTaq™ 5X PCR Master Dye Mix
5’→3’ DNA polymerase activity
No detectable 3'→5' exonuclease (proofreading) activity
Generates PCR products with 3'-dA overhangs
High yield PCR
High reproducibility
Reduced pipetting errors
Includes tracking dye for direct loading after PCR

![[RP1000] ExcelRT™ Reverse Transcriptase, 200 U/μl, 20000 U](/web/image/product.template/272/image?unique=cd0e9f9)