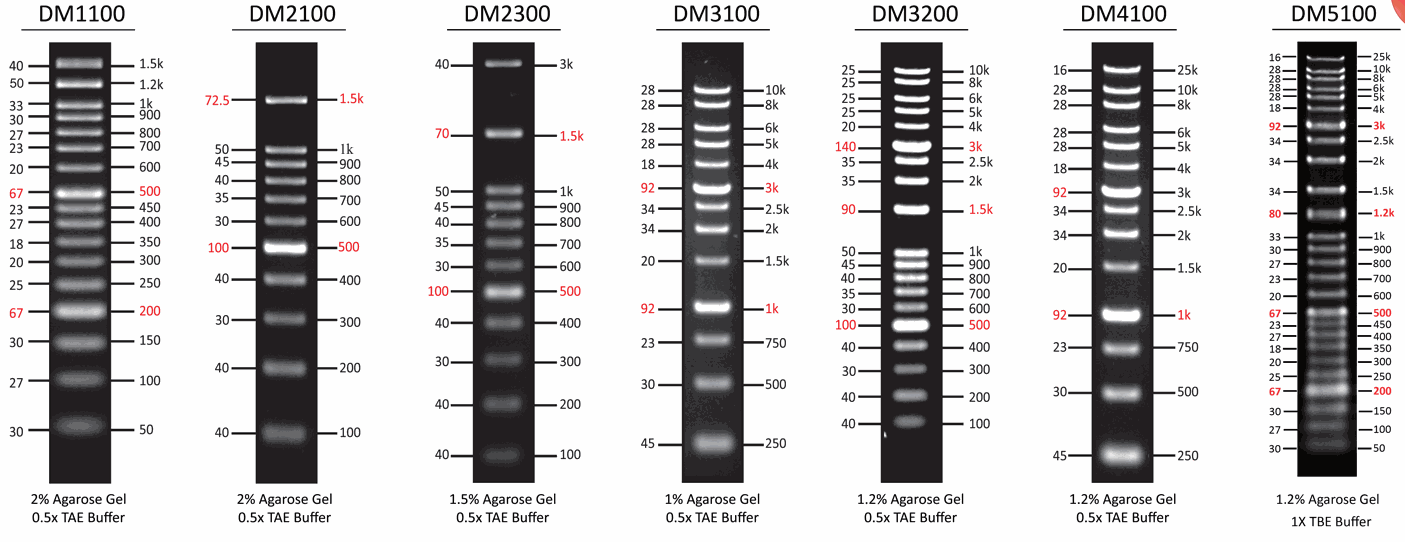
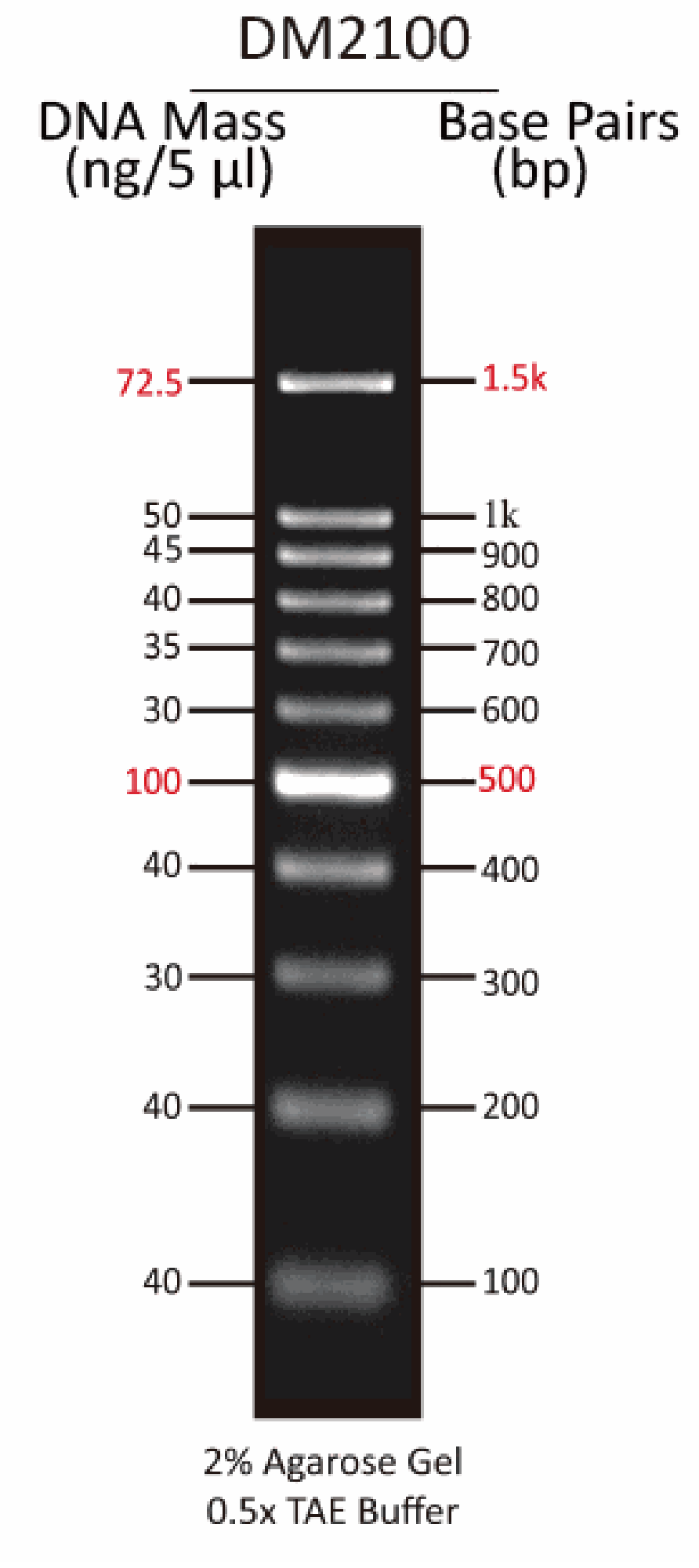
[DM2100] ExcelBand™ 100 bp DNA Ladder, 500μl
Description
The DM2100 ExcelBand™ 100 bp DNA Ladder is a ready-to-use DNA ladder, which is pre-mixed with loading dye for direct gel loading. The DNA ladder DM2100 is composed of 11 individual DNA fragments: 1.5k, 1k, 900, 800, 700, 600, 500, 400, 300, 200 and 100 bp derived from a mixture of PCR products and specifically digested plasmid DNA. This product contains two enhanced bands (1.5 kb and 500 bp) for easier reference. In addition, two tracking dyes, Xylene cyanol FF and Orange G which mimic the migration of 4,000 bp and 50 bp dsDNA during electrophoresis are also added for real time monitoring.
Features
Sharp bands
Quick reference— enhanced bands
Ready-to-use— premixed with loading dye for direct loading
Stable— room temperature storage over 6 months
Source
Phenol extracted PCR products and dsDNA digested with specific restriction enzymes, equilibrated in 10 mM Tris-HCl (pH 8.0) and 10 mM EDTA.
Range
100 ~ 1,500 bp
Concentration
52.2 µg/ 500 µl
Recommended loading volume
5 µl/ well
Storage
Room temperature for 6 months
4°C for 12 months
-20°C for 36 months
Specification
Cat. No. | DM2100 |
Series Name | ExcelBand™ |
Product Size | 500 μl |
Size Range | 100 – 1500 bp |
Band Number | 11 |
Tracking Dye | Orange G and Xylene cyanol FF |
Enhanced Band | 500 and 1500 bp |
Are the DNA markers/ladders produced by SMOBIO sufficient in quantity?
Yes, all the DNA markers of SMOBIO have been passed in the QC processes including repeated optical density measurements to ensure the quantity of total DNA.
Are the SMOBIO's DNA markers/ladders compatible to radio-labeling (for example, label DNA with T4 Polynucleotide Kinase)?
We do not recommend to do the labeling reaction directly.
The reasons are as follows:
Our DNA marker is pre-mixed with loading buffer that contains Tris-HCl, EDTA, and glycerol, may affect radio-labeling.
Our DNA marker is a mixture of PCR products and restriction enzyme digested DNA fragments. Digested dsDNA may still have phospho-group on their 5' end.
To enhance the efficiency of the labeling reaction, here are two steps suggested being done prior to the labeling reaction:
Purify DNA marker by EtOH precipitation or DNA purification kits to remove loading buffer.
Remove phospho-group by using phosphatase, ex CIAP (Calf intestinal alkaline phosphatase)
Are SMOBIO's DNA markers/ladders suitable to use in DNA PAGE?
SMOBIO's AccuBand™ series DNA markers (DM1200, DM2000, DM2200 and DM2400) are suitable for DNA PAGE.
Why DNA markers/ladders are resolved two bands at the same size when using high percentage agarose or polyacrylamide gel?
DNA fragments with identical in size are indistinguishable on agarose gels, but, on acrylamide gels, even slight differences in DNA sequence can lead to noticeably different migration rates. To provide increased intensity of DNA marker/ladder bands, multiple DNA fragments with identical in size but different in DNA sequences are used. SMOBIO's AccuBand™ series DNA markers (DM1200, DM2000, DM2200 and DM2400) are more suitable for DNA PAGE.
Are SMOBIO's DNA markers/ladders suitable to use in denaturing DNA PAGE?
SMOBIO's AccuBand™ series DNA markers (DM1200, DM2000, DM2200 and DM2400) are suitable for DNA PAGE.
SMOBIO's AccuBand™ DNA markers are not at denatured form, therefore, you need to denature DNA ladder by yourself.
Here is the protocol to denature DNA ladder:
Mix 5 μL of DNA Ladder with an equal volume of denaturing solution [95% (v/v) formamide, 10 mM EDTA (pH 8.0), 0.1% (w/v) bromophenol blue, 0.1% (w/v) xylene cyanol].
Incubate at 70°C for 5 minutes.
Electrophorese the sample in a denaturing polyacrylamide/urea gel
Enferadi A, Ownagh A, Tavassoli M. BMC Vet Res. 2025 Oct 22;21(1):627. doi: 10.1186/s12917-025-04994-4
PMCID: PMC12542350.
Ferdosi F, Jaydari A, Shams N, Khademi P. New Microbes New Infect. 2025 Nov 12;68:101666. doi: 10.1016/j.nmni.2025.101666
PMCID: PMC12666436
Patel E, Das P, Hazra S, Sharma M, Chhabra G, Gill BS, Sharma S, Kaur A, Singla D, Sandhu JS. Transgenic Res. 2025 Jun 11;34(1):29. doi: 10.1007/s11248-025-00447-8
PMCID: PMC12158852
Majidi Fard Z, Irani Mavi N, Akbari S, Erfani Y, Eghtedari S.Iran J Pharm Res. 2025 Jun 29;24(1):e160772. doi: 10.5812/ijpr-160772
PMCID: PMC12297010
Hiroshi Arai , Susumu Katsuma , Noriko Matsuda-Imai , Shiou-Ruei Lin , Maki N Inoue , Daisuke Kageyama
Elife. 2025 Apr 14:13:RP101101. doi: 10.7554/eLife.101101.
PMCID: PMC11996169
Sahar Salami , Davoud Koolivand , Omid Eini , Roghayeh Hemmati
Sci Rep. 2025 Apr 3;15(1):11377. doi: 10.1038/s41598-025-96068-6.
PMCID: PMC11965327
Ekta Patel , Piyali Das , Somak Hazra , Manveer Sharma , Gautam Chhabra , Balwinder Singh Gill , Sucheta Sharma , Ajinder Kaur , Deepak Singla , Jagdeep Singh Sandhu
PMCID: PMC12158852
Watcharapong Mitsuwan, Ratchadaporn Boripun, Phirabhat Saengsawang, Sutsiree Intongead, Sumaree Boonplu, Rawiwan Chanpakdee, Yukio Morita, Sumalee Boonmar, Napapat Rojanakun, Natnicha Suksriroj, Chollathip Ruekaewma, Titima Tenitsara
Antibiotics (Basel). 2025 Feb 25;14(3):235. doi: 10.3390/antibiotics14030235.
PMCID: PMC11939528
Quynh-Trang Do, Shun-Fen Tzeng, Chih-Yen Wang, Chih-Hsing Wu, Husam Kafeenah, Shu-Hui Chen
Commun Biol. 2025 Mar 11;8(1):357. doi: 10.1038/s42003-025-07657-0.
PMCID: PMC11897211
Prevalence of Coxiella burnetii in unpasteurized dairy products in west of Iran.
Mohammadkhanifard S, Jaydari A, Rashidian E, Shams N, Khademi P.
Parasite Epidemiol Control. 2025 Jan 23;29:e00411. doi: 10.1016/j.parepi.2025.e00411. eCollection 2025 May.
PMCID: PMC11836485
Nozawa S, Pangilinan DCJ, Dionisio GA, Watanabe K.
PLoS One. 2024 Dec 2;19(12):e0313358. doi: 10.1371/journal.pone.0313358. eCollection 2024.
PMCID: PMC11611109
Zahrakar A, Rashidian E, Jaydari A, Rahimi H.
Sci Rep. 2024 Oct 25;14(1):25271. doi: 10.1038/s41598-024-77059-5.
PMC11512005
Khademi P, Tukmechi A, Ownagh A.
New Microbes New Infect. 2024 Oct 11;62:101495. doi: 10.1016/j.nmni.2024.101495. eCollection 2024 Dec.
PMCID: PMC11533604
Yui Mizumoto-Teramura, Teru Kamogashira, Kenji Kondo, Tatsuya Yamasoba MethodsX. 2024 Jun 22:13:102818. doi: 10.1016/j.mex.2024.102818. eCollection 2024 Dec.
PMCID: PMC11267051
Saeed Shahabi, Kourosh Azizi, Qasem Asgari, Bahador Sarkari
Can J Infect Dis Med Microbiol. 2024; 2024: 4896873. Published online 2024 Mar 7. doi: 10.1155/2024/4896873
PMCID: PMC10940013
Genetic diversity of Brucella melitensis isolates from sheep and goat milk in Iran
Heidar Rahimi, Amir Tukmechi, Ehsan Rashidian Vet Res Forum. 2023; 14(12): 649–657. Published online 2023 Dec 15. doi: 10.30466/vrf.2023.1988859.3768
PMCID: PMC10759773
Ahmad Enferadi, Abdolghaffar Ownagh, Mousa Tavassoli. Vet Res Forum. 2024; 15(2): 89–95. Published online 2024 Feb 15. doi: 10.30466/vrf.2023.2000526.3855
PMCID: PMC10924296
Additions to Diatrypaceae (Xylariales): Novel Taxa and New Host Associations
Naghmeh Afshari, Omid Karimi, Antonio R. Gomes de Farias, Nakarin Suwannarach, Chitrabhanu S. Bhunjun, Xiang-Yu Zeng, Saisamorn Lumyong
J Fungi (Basel) 2023 Dec; 9(12): 1151. Published online 2023 Nov 28. doi: 10.3390/jof9121151
PMCID: PMC10744582
Shital Patil, Mohd Imran, R Sahaya Mercy Jaquline, Vidhu Aeri. ACS Omega. 2023 Aug 4;8(32):29324-29335. doi: 10.1021/acsomega.3c02543. eCollection 2023 Aug 15.
PMCID: PMC10433337
Omer Ophir, Gilad Levy, Ela Bar, Omri Kimchi Feldhorn, May Rokach, Galit Elad Sfadia, Boaz Barak. Biomedicines. 2023 Aug 15;11(8):2273. doi: 10.3390/biomedicines11082273.
PMCID: PMC10452363
Saeed Shahabi, Kourosh Azizi, Qasem Asgari, Bahador Sarkari. J Trop Med. 2023 Feb 10;2023:5965340. doi: 10.1155/2023/5965340. eCollection 2023.
PMCID: PMC9937756
Noraikim Mohd Hanafiah, Acga Cheng, Phaik-Eem Lim, Gomathy Sethuraman, Nurul Amalina Mohd Zain, Niranjan Baisakh, Muhamad Shakirin Mispan
Life (Basel). 2022 Oct; 12(10): 1542. Published online 2022 Oct 4. doi: 10.3390/life12101542
PMCID: PMC9604669
Ferdinando Bonfiglio, Vito Alessandro Lasorsa, Sueva Cantalupo, Giuseppe D'Alterio,a,d Vincenzo Aievola, Angelo Boccia, Martina Ardito, Simone Furini, Alessandra Renieri, Martina Morini, Sabine Stainczyk, Frank Westermann, Giovanni Paolella, Alessandra Eva, Achille Iolascon, Mario Capasso
eBioMedicine. 2023 Jan; 87: 104395. Published online 2022 Dec 6. doi: 10.1016/ j.ebiom.2022.104395
PMCID: PMC9732128
Muzaffar Mosquill, Syafinaz Amin Nordin, Mohamad Ridhuan Mohd Ali, Narcisse Mary Sither Joseph, MethodsX. 2022 Jul 30;9:101804. doi: 10.1016/j.mex.2022.101804. eCollection 2022.
PMCID: PMC9382312
Amin Khoshbayan, Rezvan Golmoradi Zadeh, Majid Taati Moghadam, Shiva Mirkalantari, Atieh Darbandi, Ann Clin Microbiol Antimicrob. 2022 Aug 4;21(1):35. doi: 10.1186/s12941-022-00526-2.
PMCID: PMC9351160
Genomic Detection of Brucella Abortus in Milk obtained from Farms in Lorestan Province using PCR Method
Rahim Tahmasebi, Amin Jaydari, Nemat Shams, Heidar Rahimi, Iran J Med Microbiol September - October 2022, Issue 5 , 16(5): 479-484.
Characterization of Staphylococcus aureus isolates from pastry samples by rep-PCR and phage typing
Mohammad Reza Arabestani, Farideh Kamarehei, Mahya Dini, Farid Aziz Jalilian, Abbas Moradi, Leili Shokoohizadeh, Iran J Microbiol. 2022 Feb;14(1):76-83. doi: 10.18502/ijm.v14i1.8806.
PMCID: PMC9085548
Development of SCAR Markers to Identify Medicinal Cultivars of Paeonia lactiflora
Yuina Yoshie, Hirokazu Ando, Takayuki Tamura, Kozo Fukuda, Motoko Igarashi, Atsuyuki Hishida, Nobuo Kawahara, Yohei Sasaki, Biol Pharm Bull
. 2022;45(3):292-300. doi: 10.1248/bpb.b21-00810.
PMID: 35228395
Functional kupffer cells migrate to the liver from the intraperitoneal cavity
Wen-Ling Lin, Mizuki Mizobuchi, Mina Kawahigashi, Otoki Nakahashi, Yuuki Maekawa, Takashi Sakai, Biochem Biophys Rep. 2021 Aug 17;27:101103. doi: 10.1016/j.bbrep.2021.101103. eCollection 2021 Sep.
PMCID: PMC8379421
Phylogenetic and molecular analysis based on genes 16S-rRNA, OMPA and POMP to identify Chlamydia abortus infection occurrence at the milk samples of goats and sheep in west Azerbaijan of Iran
Fariba Taheri, Abdulghaffar Ownagh, Karim Mardani, Iran J Microbiol. 2021 Aug;13(4):480-487. doi: 10.18502/ijm.v13i4.6972.
PMCID: PMC8421589
Eggshell membrane for DNA sexing of the endangered Maleo ( Macrocephalon maleo)
Pramana Yuda, Conceptualization, Formal Analysis, Methodology, Writing – Original Draft Preparation, Writing – Review & Editinga,1 and Andie Wijaya Saputra, Data Curation, Investigation, Project Administration, Writing – Original Draft Preparation1, Version 4. F1000Res. 2020; 9: 599. Published online 2021 Jan 28. doi: 10.12688/f1000research.23712.4
PMCID: PMC7836084
Chih-Ying Lin, Lih-Yuan Lin, PLoS One. 2018; 13(1): e0191971. Published online 2018 Jan 30. doi: 10.1371/journal.pone.0191971
PMCID: PMC5790263
Katherine Domb, Danielle Keidar, Beery Yaakov, Vadim Khasdan, Khalil Kashkush, BMC Plant Biol. 2017; 17: 175. Published online 2017 Oct 27. doi: 10.1186/s12870-017-1134-z
PMCID: PMC5659041

FluoroVue™ Nucleic Acid Gel Stain
Excellent for in-gel staining
Sensitivity up to 0.14 ng DNA or 1 ng total RNA
A safer alternative to EtBr
Suitable to blue or UV light

FluoroStain™ DNA Fluorescent Staining Dye
Excellent for post staining
Sensitivity up to 0.04 ng DNA
A safe alternative to EtBr
Suitable for blue or UV light

FluoroDye™ DNA Fluorescent Loading Dye
Excellent for premix with DNA sample
Sensitivity up to 0.14 ng DNA
Safety dye
Convenience - monitor the electrophoresis in real-time

![[DM2100] ExcelBand™ 100 bp DNA Ladder, 500μl](/web/image/product.template/222/image?unique=3244435)
![[DM2100] ExcelBand™ 100 bp DNA Ladder, 500μl](/website/image/product.template/222/image/90x90)