Description
The EzRNA™ T7 High Yield RNA Synthesis Kit is a user-friendly product for enzymatic RNA production. The enzyme mix contains adequate amount of T7 RNA polymerase, pyrophosphatase, and RNase inhibitors for in vitro transcription (IVT). Along with 10X Transcription Buffer and NTP Premix, users can swiftly assemble IVT reactions without compromising RNA yield. The EzRNA™ T7 High Yield RNA Synthesis Kit allows for the attainment of approximately up to 150 µg RNA yield within 2 hours at 37°C.
Features
High yield
Versatile- suitable for short and long transcripts
NTP premixed- Minimal pipetting and setup time
Compatible with CleanCap® Reagent AG
Lithium chloride included for RNA purification
Application
Generation of RNA from T7 promoter-driven DNA sequences
Suitable for subsequent cap-0 and cap-1 modification
Storage
-20°C for 12 months
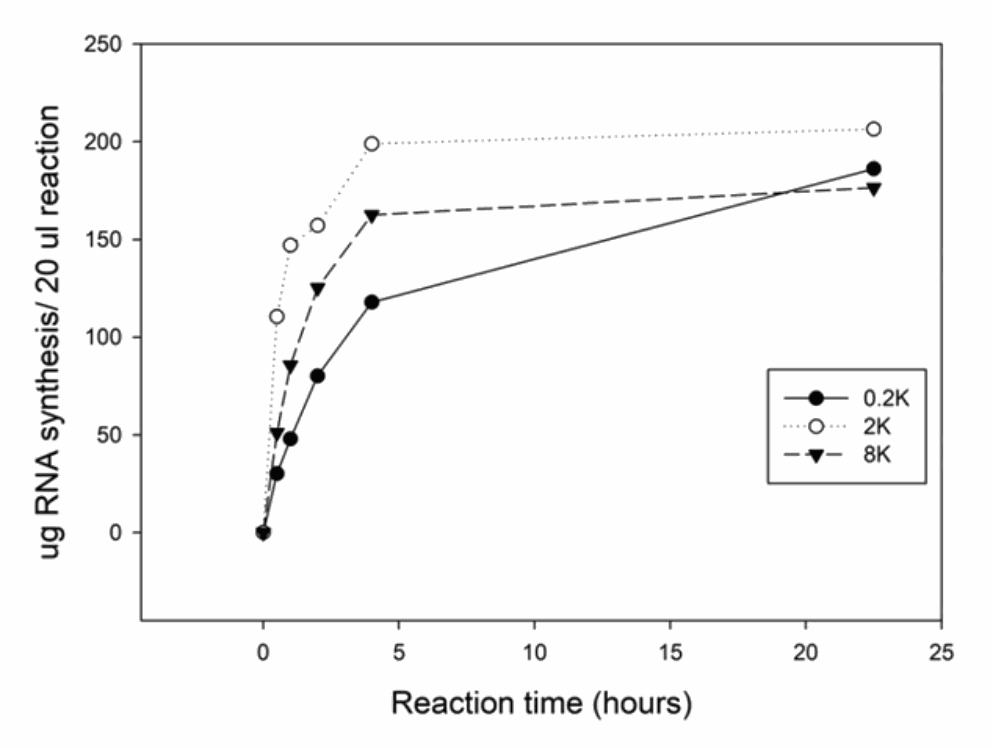
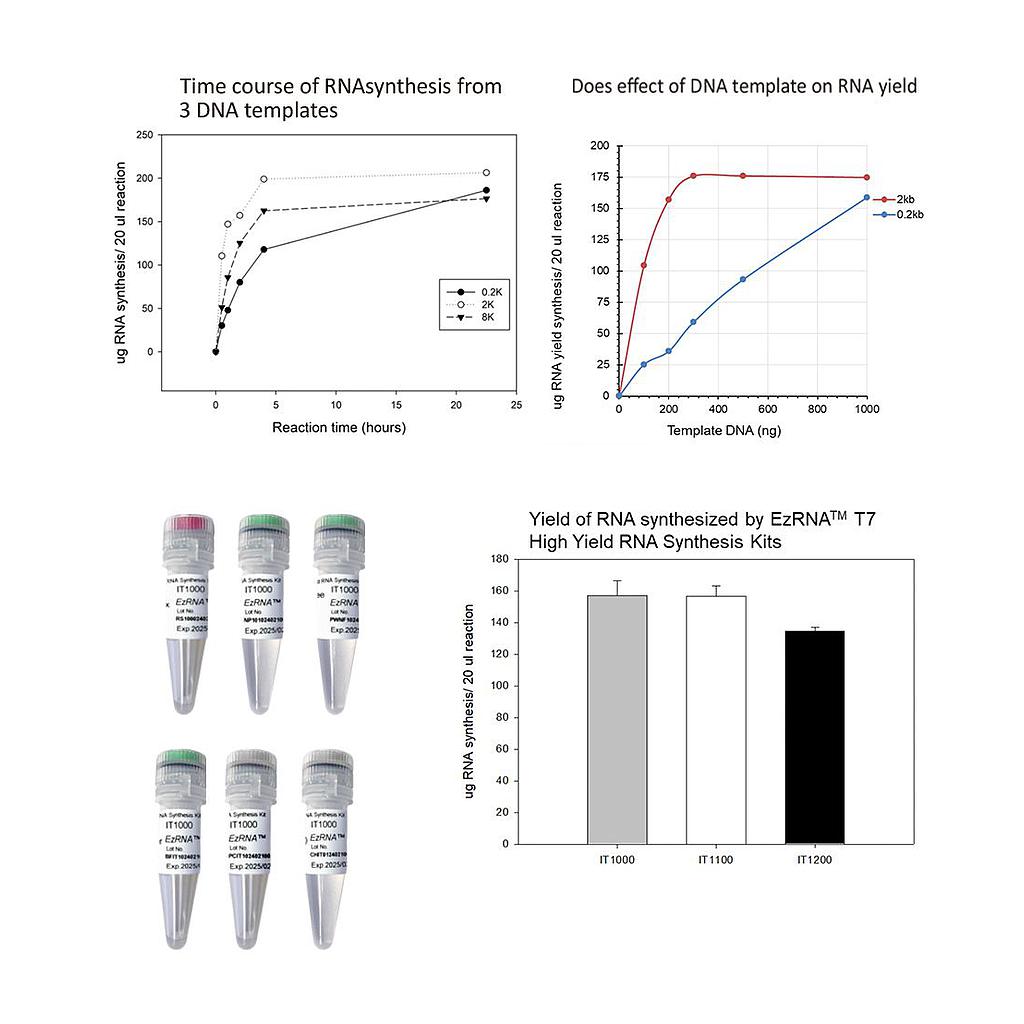
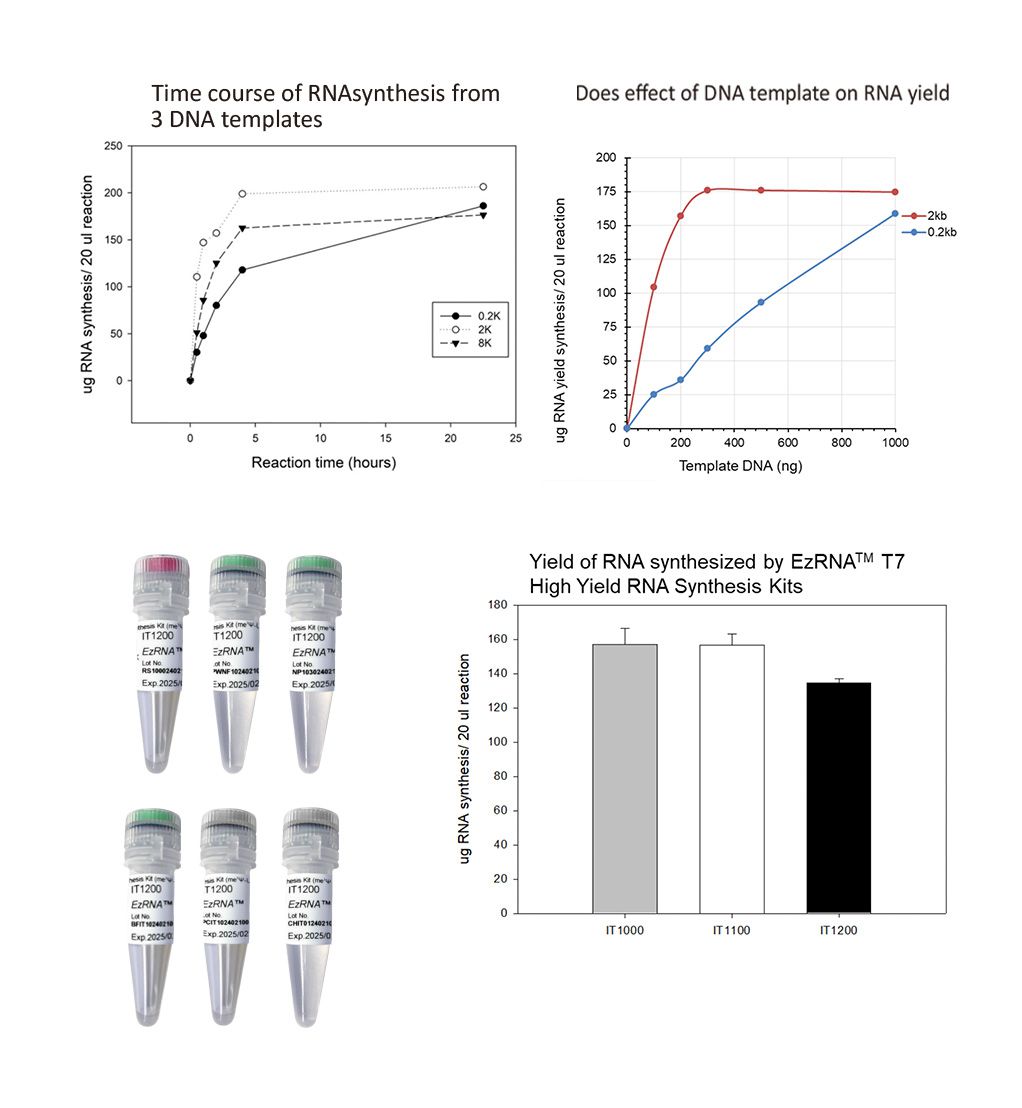
Time course of RNA synthesis from 3 DNA templates
The 0.2k, 2k and 8k RNA transcripts were generated using the SMOBIO EzRNA™T7 High Yield RNA Synthesis Kit, the reaction containing 1 µg DNA template was incubated at 37°C for indicated reaction time.
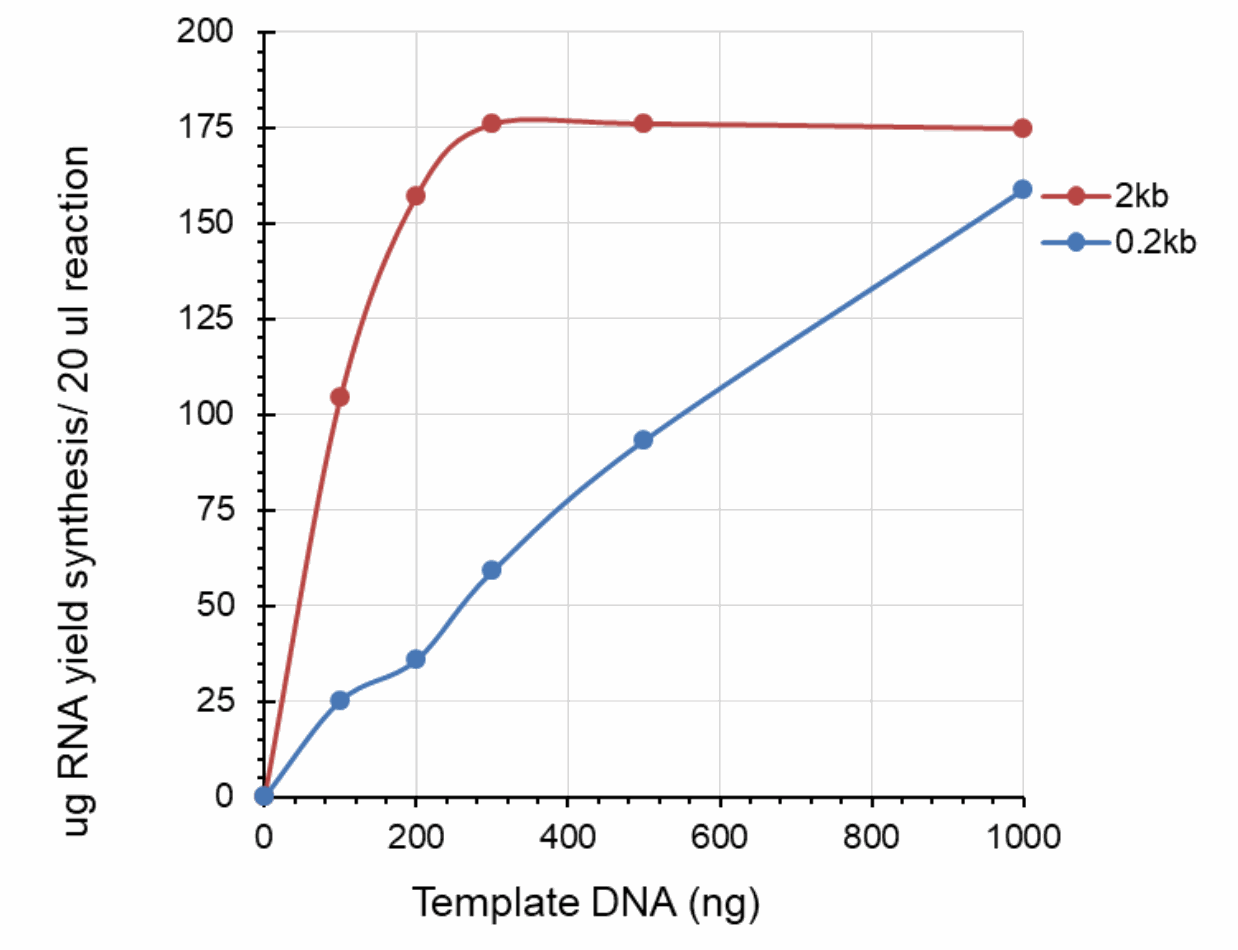
Dose effect of DNA template on RNA yield
The 0.2k and 2k RNA transcripts were generated using the SMOBIO EzRNA™T7 High Yield RNA Synthesis Kit , the reaction containing indicated amounts of DNA template was incubated at 37°C for 2 hours.
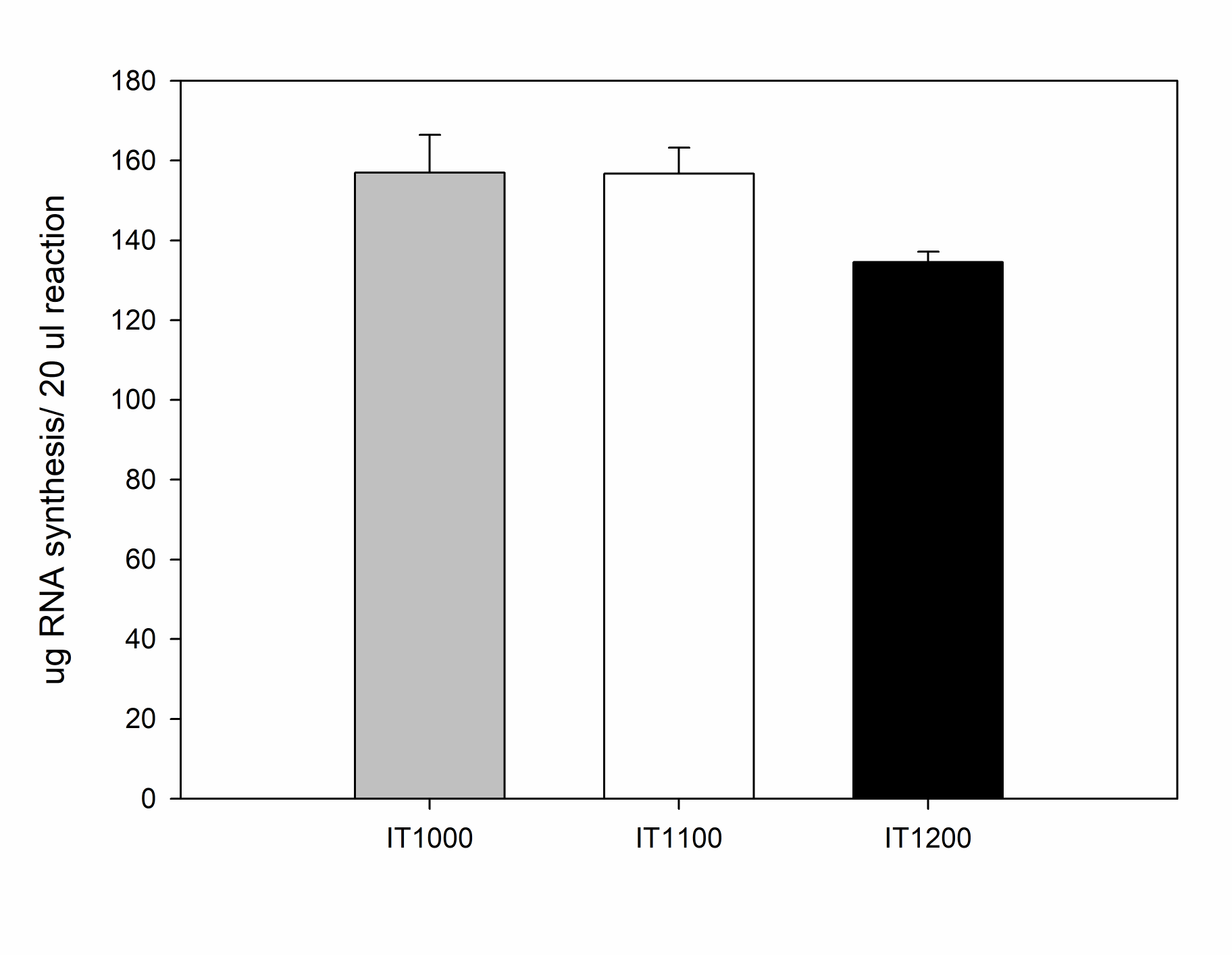
Yield of RNA synthesized by EzRNA™ T7 High Yeield RNA Synthesis Kits
Contents
Components | Volume |
T7 RNA Polymerase Mix | 100 μl |
10X T7 Reaction Buffer | 100 μl |
NTP Premix (25 mM each) | 400 μl |
Control DNA (0.5 μg/μl) | 10 μl |
Lithium Chloride (7.5M) | 1 ml |
Nuclease-Free Water | 1 ml |
Storage
-20°C for 12 months
What is the length of the Control DNA? How to use the Control DNA in IVT Reaction?
1. The Control DNA is approximately 2,110 base pairs in length and is provided as linearized DNA, which can be used directly in the IVT reaction without any additional processing. The RNA produced in the IVT reaction will typically have the same length as the DNA template, around 2,110 nucleotides.
2. The concentration of the Control DNA is 0.5 μg/μL. For a standard IVT reaction, we recommend using 1 μg of DNA (approximately 2 μL). However, if you need to use the Control DNA more frequently, you can reduce the amount used per reaction (e.g., 0.5 μg) to extend the number of uses, while still maintaining experimental reliability.
What type of DNA template is suitable for performing in vitro transcription?
A. Double stranded linear DNA with blunt or 5'-protruding ends is suitable template for in vitro transcription reaction. Linearized plasmid DNA, PCR products or cDNA can be used as templates for transcription if they contain a double-stranded T7 promoter region in the correct orientation.
B. The sequence of T7 promoter: 5'-taatacgactcactataG*-3'. G* will be the first base of the RNA transcript. The synthesis of the sense or antisense RNA transcript depends on the orientation of the T7 promoter with respect to target sequence.
The target sequence must be placed downstream of the T7 promoter for sense RNA and must be inverted for antisense RNA transcription.
What measures can be taken to prevent RNase contamination during an IVT reaction?
Every component within the IVT kit has undergone testing to confirm the absence of contaminating ribonuclease activities. Nevertheless, the creation of an RNase-free working environment and the use of RNase-free solutions are essential prerequisites for achieving successful in vitro transcription.
General recommendations to avoid RNase contamination are:
A. Maintain a separate area, dedicated pipettors and reagents for RNA work.
B. Wear gloves when handling RNA and reagents for work with RNA. Change gloves frequently.
C. Use sterile RNase-free plastic tubes and pipette tips.
D. Treat water and all solutions used for RNA purification and handling with DEPC-H2O. Add DEPC to a final concentration of 0.1% (v/v), incubate overnight at room temperature and autoclave.
E. Keep all kit components sealed when not in use and all tubes tightly closed during transcription reaction.
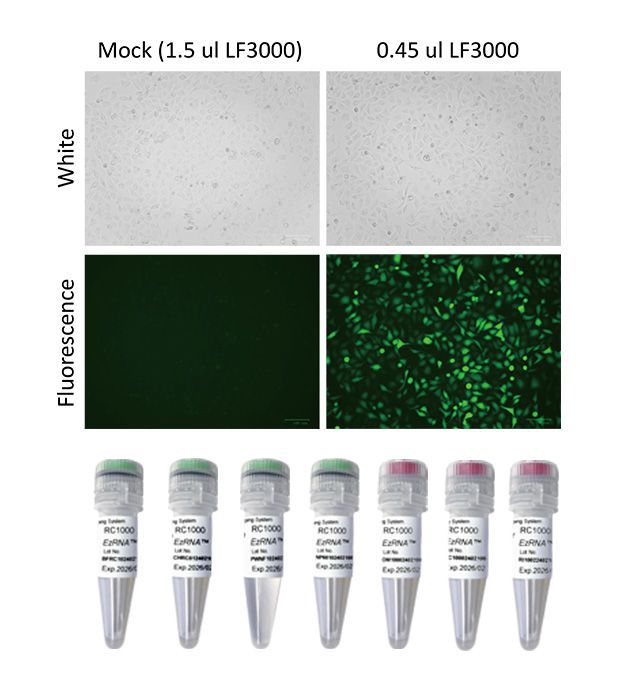
If in vitro-transcribed RNA can be directly used for cell transfection assay or other in vivo studies?
In vitro-transcribed (IVT) RNA is generally not recommended for cell transfection or in vivo studies, certain considerations need to be addressed before its direct use.
A. RNA modifications: Some modifications, such as a 5' cap structure and a poly(A) tail, are often added to IVT RNA to enhance stability and translational efficiency. The absence of these modifications in IVT RNA may result in reduced expression levels and rapid degradation in cells.
B. Purity and integrity: The quality of IVT RNA is crucial for successful transfection or in vivo studies. Contaminants such as template DNA, short RNA fragments, or unincorporated nucleotides can affect transfection efficiency and cell viability. Therefore, it's important to purify the IVT RNA to remove these impurities.
C. Transfection reagents: The choice of transfection reagent is critical for the successful delivery of IVT RNA into cells. Not all transfection reagents are suitable for RNA transfection, and optimization of the transfection protocol may be necessary for specific cell types.
What might be the cause of a lower transcript yield than expected?
If the yield of the control group is normal while the yield obtained from experimental group is low, there may be quality problems in the template of the experimental group. It is recommended to try the following solutions:
A.The DNA template may contain components that inhibit the IVT reaction. Please repurify the DNA template. The template should be RNase-free and at high concentration
B.Determine template quantity and its integrity
C.Extend transcription reaction time
D.Increase template input
How can a low IVT yield for short RNA transcripts be addressed?
If the transcription fragment is too short, the reaction will be inhibited. When the transcription length is less than 500 nt, please prolong the reaction time and increase the amount of template input. It is recommended to use 2 μg of template. For the transcription of super-short fragments e.g. 100 nt, it is recommended to add 2 μg of template to reaction mixture for at least 6 hours, and 16 hours of overnight transcription for maximum yield.
After in vitro transcription, why is a turbid appearance observed on the reaction mixture?
The transcription mixture typically starts to become turbid approximately 15 minutes after setting up the reaction. This turbidity, or increase in viscosity, indicates RNA transcription occurring and eventual precipitation of RNA out of the reaction mixture.
After in vitro transcription, how do I select an appropriate purification method for my RNA transcript?
Lithium chloride (LiCl) precipitation is efficient in eliminating unbound NTPs and proteins.
However, if the RNA size is below 300 nt or the concentration is less than 0.1 μg/μl, effective RNA recovery becomes challenging. In such cases, consider purifying the RNA through spin column or ethanol precipitation following phenol-chloroform extraction. RNA purified using these methods is suitable for downstream applications such as cell transfection, electroporation, and microinjection experiments.
Can the EzRNATMT7 High Yield RNA Synthesis Kit be used in conjunction with Cap analog for RNA capping?
The compatibility with Cap analog depends on the specific type of the Cap analog utilized. When CleanCap® Reagent AG is employed, the RNA yield generated by the EzRNATM T7 High Yield RNA Synthesis Kit is minimally affected and thus it is compatible with EzRNATM T7 High Yield RNA Synthesis Kit.
However, when co-transcribing with conventional Cap analogs such as ARCA (anti-reverse cap analog), the RNA yield may decrease. This reduction becomes more significant with higher concentrations of ARCA. The decrease in yield is primarily attributed to the competition between ARCA and GTP (guanosine triphosphate) for incorporation into the RNA transcript, leading to incomplete transcripts and an overall reduction in RNA yield.
Thus, it is advisable not to combine the EzRNATM T7 High Yield RNA Synthesis Kit with conventional Cap analogs but instead to use CleanCap® Reagent AG or enzymatic capping method for RNA capping.
Is it necessary to remove DNA template after IVT reaction?
Yes, it is generally necessary to remove the DNA template after an IVT reaction, especially if the RNA product is intended for downstream applications where DNA contamination can interfere with the results or compromise the experiment's integrity. Removing the DNA template after an IVT reaction is essential to maintain the integrity, specificity, and purity of the RNA for downstream applications. It helps ensure accurate and reliable results in molecular biology experiments.
What methods are commonly used to remove DNA template after IVT reaction?
A. DNase treatment: Add DNase I into the IVT reaction mixture to degrade any residual DNA template after IVT reaction.
DNase I is selective in cleaving both single-stranded and double-stranded DNA while preserving the integrity of RNA.
Subsequently, apply heat to inactivate DNase I, ensuring its inactivation and the preservation of RNA integrity.
B. Lithium Chloride (LiCl) precipitation: LiCl precipitation is a commonly used method to selectively precipitate RNA while leaving DNA and other contaminants in solution.
However, it is important to note that LiCl precipitation may not be suitable for all RNA samples (e.g. RNA size < 300 nt), and alternative purification methods may be preferred based on specific requirements.
What's the reason for using modified nucleotides such as pseudouridine-5'-triphosphate (Ψ-TP) and N1-methylpseudouridine-5'-triphosphate (me1Ψ-TP) in IVT RNA synthesis?
The use of modified nucleotides in IVT RNA synthesis can improve the stability, translational efficiency, and reduce immunogenicity of RNA molecules, making them more suitable for various experimental and therapeutic applications.
A. Enhanced stability: Modified nucleotides can enhance the stability of the resulting RNA molecules. For instance, 2'-O-methylated nucleotides can increase RNA stability by protecting against enzymatic degradation by nucleases.
B. Improved translation efficiency: Certain modifications, such as pseudouridine (Ψ), can enhance translation efficiency by facilitating ribosome binding and elongation during protein synthesis.
C. Reduced immunogenicity: Unmodified RNA molecules may elicit an immune response when introduced into cells or organisms. Modified nucleotides, such as N1-methyl pseudouridine, can reduce the immunogenicity of IVT RNA by minimizing recognition by cellular innate immune sensors.
How is the poly(A) tail added to IVT RNA?
For in vitro-transcribed RNA, the poly(A) tail can be encoded in the DNA template by using an appropriately tailed PCR primer, or enzymatically added to the RNA by Poly(A) Polymerase in a separate step following IVT reaction.
After in vitro transcription, how to quantify my RNA transcript?
To assess RNA concentration most conveniently, utilize ultraviolet light absorption at a wavelength of 260 nm. Dilute a portion of the reaction at a 1:300 ratio to achieve an absorbance reading within the linear range of a spectrophotometer.
For single-stranded RNA, when A260 = 1, RNA concentration is 40 μg/mL. The RNA yield can be determined using the formula: A260 × 300 (dilution factor) × 40 = __ μg/mL RNA.
It's important to note that unincorporated nucleotides and template DNA in the mixture may interfere with the reading. Therefore, for precise quantification, it is advisable to remove the template and nucleotides from the transcription mixture.
Before setting up the reactions, please gently mix the components with a pipette and centrifuge briefly to collect at the bottom of the tube.
Please prepare the reaction at room temperature: When preparing the reaction mixture, except for the T7 RNA Polymerase Mix, which should be temporarily stored on ice, other components should be kept at room temperature, and the reaction should be prepared at room temperature.
Please include each reaction component following this sequence: Water → Reaction Buffer → NTP Premix → DNA template → T7 RNA Polymerase. Ensure that the template and enzyme are added last.
Make sure reactions are thoroughly mixed prior to subject to 37℃.
To avoid volatilization of the reaction solution due to prolonged transcription, we recommend incubating the IVT reactions in a PCR machine or in a dry air incubator.
[RC1000] EzRNA™ RNA Capping System, 50 RXN
Application
Generation of 5’Cap-0 (m7Gppp) and Cap-1 (m7GpppNm-) RNA by enzymatic reaction
mRNA synthesis for in vitro translation
Gene expression studies
mRNA vaccine development and therapeutics
[IT1100] EzRNA™T7 High Yield RNA Synthesis Kit (Ψ-UTP), 50 RXN
Application
Generation of RNA from T7 promoter-driven DNA sequences
Suitable for subsequent cap-0 and cap-1 modification
[IT1200] EzRNA™ T7 High Yield RNA Synthesis Kit (me1Ψ-UTP), 50 RXN
Application
Generation of RNA from T7 promoter-driven DNA sequences
Suitable for subsequent cap-0 and cap-1 modification
FluoroVue™ Nucleic Acid Gel Stain
Excellent for in-gel staining
Sensitivity up to 0.14 ng (DNA) or or 1 ng (total RNA)
A safe alternative to EtBr
Suitable for blue or UV light
Excellent for in-gel staining
Sensitivity up to 0.14 ng (DNA) or or 1 ng (total RNA)
A safe alternative to EtBr
Suitable for blue or UV light