2. mRNA stock
mRNA manufacturing could be separated into 3 parts, as listed in following sections:
1. In Vitro Transcription, IVT
Aside from DNA raw material, there are a few of enzymes and nucleotides involved in the IVT reaction described below:
:
(1)
T7 RNA polymerase
:
It can recognize and bind promotor region, and then start to proceed transcribe RNA strand based on DNA template.
(2)
RNase inhibitor
:
It’s the element that specifically block RNase A, which results in preserving RNA quality during IVT reaction.
(3)
Nucleotide raw materials
:
They are the crucial RNA building block that including ATP, UTP, CTP, and GTP. Additionally, uridine moiety has been substituted by N1-methylpeudouridine in Pfizer/BioNTech and Moderna COVID-19 vaccine, to enhance RNA stability and translation efficiency.
(4)
Inorganic Pyrophosphatase (PPase)
:
It could increase RNA production by hydrolyzing pyrophosphate, which reduces the amount of pyrophosphate and thus promotes the reaction rate of RNA synthesis.
Technical Demo 1:
SMOBIO has built up IVT RNA product line since 2015. We have the ability to produce RNA ranging from 200 base to 6 kilo-base long, and assemble them as RNA marker. The workflow of RNA marker production is shown below:
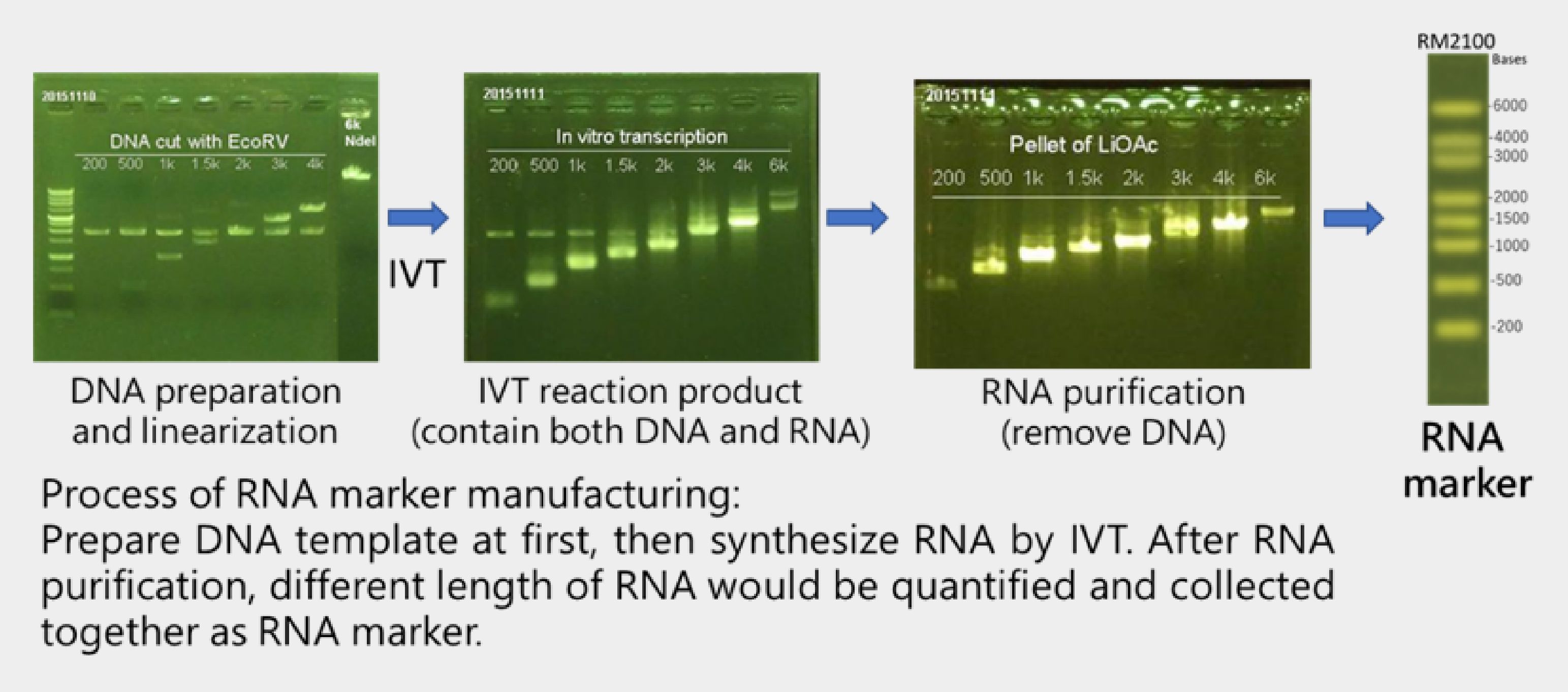
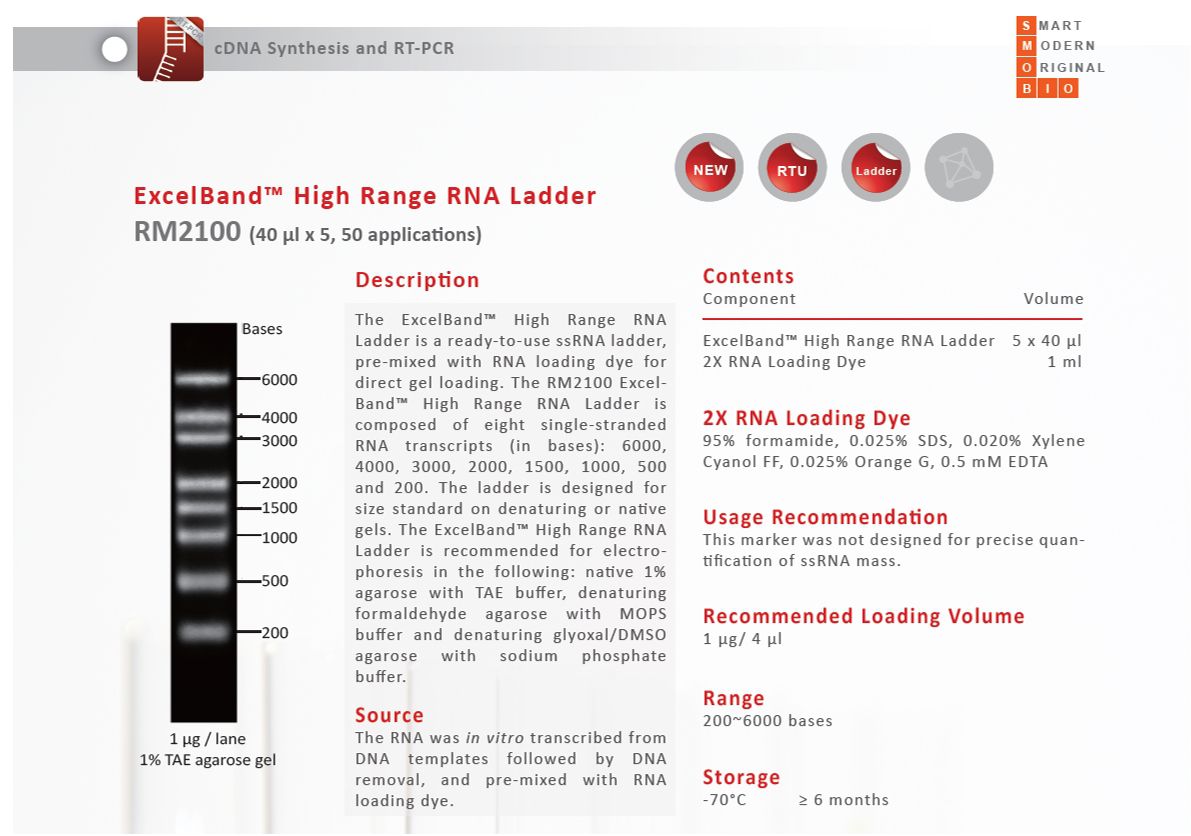
The RNA marker datasheet showed above were derived from 2016 SMOBIO catalog.
Technical Demo 2:
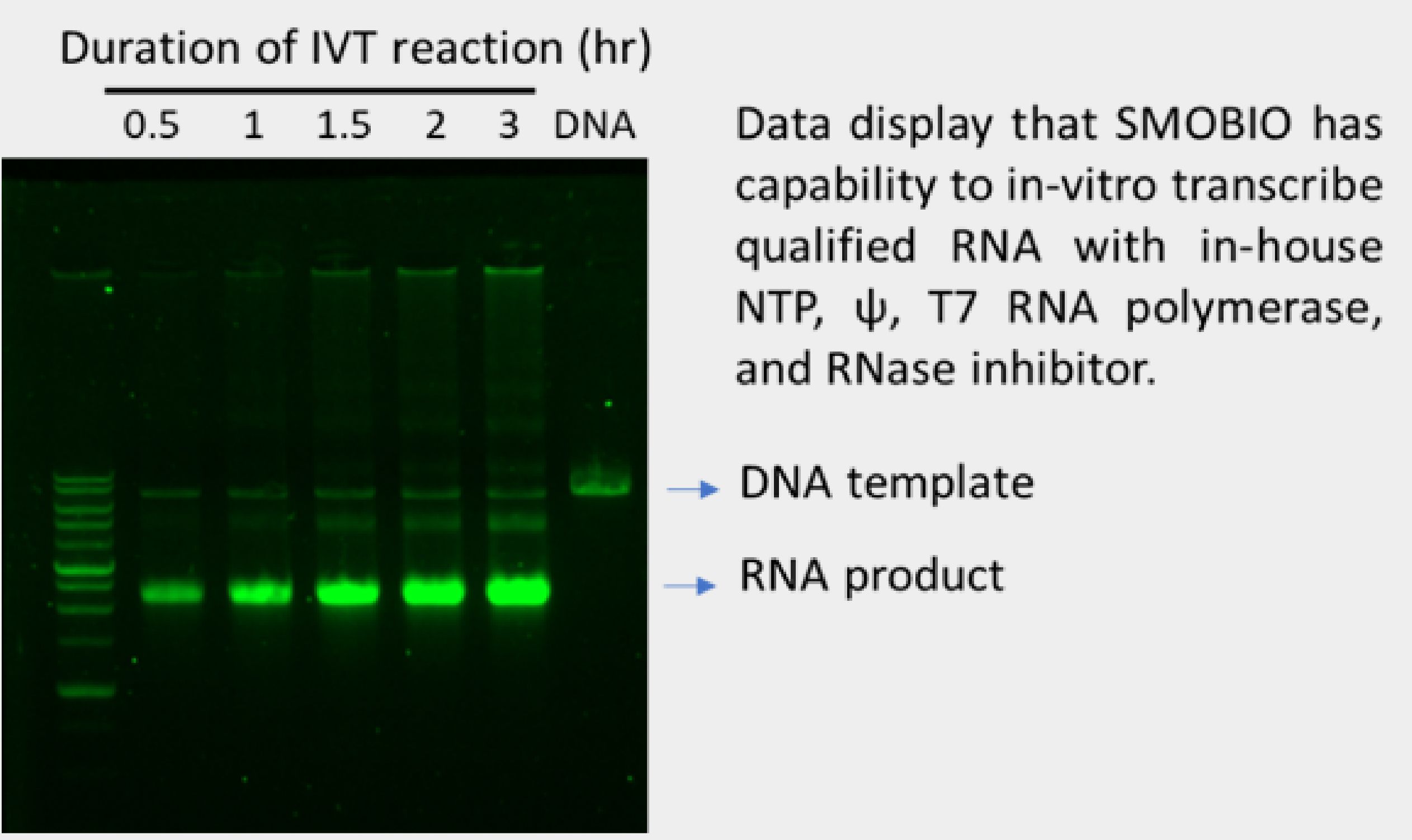
With our in-house production of T7 polymerase, nucleotide including pseudouridine triphosphate, RNase inhibitor, and DNA construct based on Pfizer/BNT mRNA vaccine framework, SMOBIO has successfully synthesized modified mRNA encoding Covid-19 Spike as shown below:
2. RNA modification
Three main kinds of modification including capping, poly(A) tailing, and cap methylation are described in following paragraph:
(1)
Capping
:
Cap of RNA is the vital structure in eukaryotic cells, which can bind with elongation factor 4E (eIF4E) to initiate translation process. Also, capped structures are able to protect 5’end RNA from degradation thereby elevating RNA stability. There are two ways for capping. One is the single step method used by Pfizer/BioNTech, where incubating cap analog along with RNA synthesis so that cap would be appended in the same reaction. The other one is two-step method taken by Moderna, an additional step would be applied after the IVT procedure, where adding 7-methylguanosine cap (Cap 0) to 5’end of RNA by vaccinia capping enzyme D1 and D12 subunits. It usually comes with 2’O-methyltransferase reaction.
.
(2)
Methylation
:
Aside from capping, methylation at 2’position of cap-1 ribose (that is, the first nucleotide followed cap structure) would promote RNA stability as well.
(3) Poly(A) tailing:Poly(A) tail is the essential structure of mRNA and related to RNA stability as well as translation efficiency. There are two ways for poly(A) tailing as mentioned. One is built in DNA template then to create poly(A) with IVT reaction, while the other is appending after RNA synthesis.
Three main kinds of modification including capping, poly(A) tailing, and cap methylation are described in following paragraph:
(1)
Capping
:
Cap of RNA is the vital structure in eukaryotic cells, which can bind with elongation factor 4E (eIF4E) to initiate translation process. Also, capped structures are able to protect 5’end RNA from degradation thereby elevating RNA stability. There are two ways for capping. One is the single step method used by Pfizer/BioNTech, where incubating cap analog along with RNA synthesis so that cap would be appended in the same reaction. The other one is two-step method taken by Moderna, an additional step would be applied after the IVT procedure, where adding 7-methylguanosine cap (Cap 0) to 5’end of RNA by vaccinia capping enzyme D1 and D12 subunits. It usually comes with 2’O-methyltransferase reaction.
.
(2) Methylation : Aside from capping, methylation at 2’position of cap-1 ribose (that is, the first nucleotide followed cap structure) would promote RNA stability as well.
(3) Poly(A) tailing:Poly(A) tail is the essential structure of mRNA and related to RNA stability as well as translation efficiency. There are two ways for poly(A) tailing as mentioned. One is built in DNA template then to create poly(A) with IVT reaction, while the other is appending after RNA synthesis.
3. RNA purification
Generally, RNA purification includes steps for DNA template removal and ultimate RNA clean-up.
(1)
DNA template removal
:
This step is to prevent any DNA residual from RNA stock solution. Regular method is DNase reaction for DNA digestion.
(2) RNA clean-up
After DNA depletion, RNA product is required to isolate from the solution. Modern techniques cover from applying oligo-T magnetic bead, oligo-T chromatography column, lithium salt precipitation, to tangential flow filtration (TFF). There are a number of enzymes and raw materials in RNA manufacturing that need to be removed before further serving as the vaccine. Like DNA, DNase, proteinase K. Even those byproducts such as partial/full dsRNA and DNA-RNA hybrid may be recognized by specific cellular receptors, which would trigger innate immunity as well as induce alpha-interferon expression, resulting in reduction of protein translation. The current regular isolation technique is applying oligo-T purification. Given the specific pairing of adenosine (A) and thymidine (T), oligo-T can capture poly(A) under neutral or high salt environment, followed by elution of poly(A)-containing RNA from low salt condition. The RNA recovery rate could be over 90 percent.
Quality control of mRNA involves monitoring its quantity, purity and impurities (e.g. incomplete mRNA or dsRNA), as well as sequence accuracy, capping rate, poly(A) length and so on during the process. RNA quantity is usually measured by absorbance of 260 nm and 280 nm, whereas sequence accuracy can be validated by reverse transcription and subsequent DNA sequencing. Mass spectrometry analysis for overall and individual molecular weight is the major method for assessing capping rate, poly(A) length, and nucleoside modification.
