[PM1600] ExcelBand™ All Blue Regular Range Plus Protein Marker (9-180 kDa), 250 μl x 2
Description
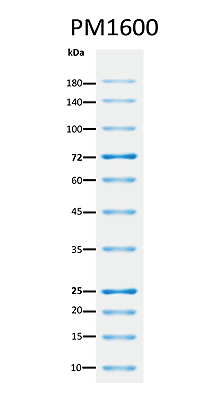
The PM1600 ExcelBand™ All Blue Regular Range Plus Protein Marker is a blue protein standard with 11 pre-stained proteins covering a wide range of molecular weights from 10 to 180 kDa in Tris-Glycine buffer (9 to 170 kDa in Bis-Tris (MOPS) buffer and Bis-Tris (MES) buffer). Proteins are covalently coupled with a blue chromophore, and two reference bands (at 25 kDa and 72 kDa, respectively) are enhanced in intensity when separated on SDS-PAGE (Tris-Glycine buffer).
The PM1600 ExcelBand™ All Blue Regular Range Plus Protein Marker is designed for monitoring protein separation during SDS-polyacrylamide gel electrophoresis, verification of Western transfer efficiency on membranes (PVDF, nylon, or nitrocellulose) and for approximating the size of proteins.
Features
Ready-to-use — Premixed with a loading buffer for direct loading, no need to boil.
Two enhanced bands — 72 kDa and 25 kDa
Contents
Approximately 0.1~0.5 mg/ml of each protein in the buffer (20 mM Tris-phosphate (pH 7.5 at 25°C), 2% SDS, 0.2 mM DTT, 3.6 M urea, and 15% (v/v) glycerol).
Quality Control
Under suggested conditions, PM1600 ExcelBand™ All Blue Regular Range Plus Protein Marker resolves 11 major bands in 15% SDS-PAGE (Tris-Glycine buffer, MOPS, and MES buffer) and after Western blotting to nitrocellulose membrane.
Storage
4°C for 3 months
-20°C for long term storage
Specification
Cat. No. | PM1600 |
Series Name | ExcelBand™ |
Product Size | 250 μl x 2 |
MW Range | 10 – 180 kDa |
Band Number | 11 |
Band Color | All blue |
Enhanced Bands | 25, 72 kDa |
Manual
Manual_PM1600_ExcelBand™ All Blue Regular Range Plus Protein Marker (2023 ver. 2.4.0)
SDS
Migration patterns and approximate MWs (kDa)
Why are there contrasting results in molecular weights after using different brands of protein markers?
A. Different proteins even with similar molecular weights would exhibit apparent disparity from the resulting SDS PAGE due to the difference in the composition of the protein’s amino acids (e.g. gelatin). The reason for the disparity is due to the amino acids composition that affects the binding of the protein and SDS. Therefore, we can say that protein marker is a handy tool to estimate molecular weight, but there is no absolute molecular weight standard.
B. While running SDS-PAGE, protein mobility can be affected by the composition of the buffer used, gel percentage, the voltage used, running time, as well as if there is a pre-run.
C. Another recommendation for high molecular weight proteins is to prolong the running time to clarify the relative location of bands.
Protein marker Retention Period: Mentioned -20°C and over 2 years. Is it available for 30 months or 36 months? Have you tested this period?
Yes, we have tested our PM2700. The results showed that the PM2700 is stable at -20℃ for at least two years. It has also shown strong performance for more than 36 months under our careful storage. However, we must only suggest a 2 year retention period for the following reasons: There may be a variation in the environment in storage, and improper use may lead to accumulated damage to the proteins and therefore reduce its retention period.
How many times of freezing and thawing are available for protein markers? If it uses 5 μL per load, would the total usage quantity be 50 times x 2 (250 μL x 2 tube)?
Yes, 100 uses (5 μL each time) can be expected if freezing and thawing are conducted carefully and properly at the appropriate temperature. Before each use, make sure the protein marker is thoroughly thawed.
Do you have data comparison for protein molecular weight’s precision with other protein markers?
Yes. Usually, pre-stained marker is written on “estimated molecular weight” for caution. It is known that the analysis of protein size by an SDS-PAGE is only for “estimation” because of the intrinsic variation of amino acid composition in all proteins including stained and non-stained ones. For example, a protein which is highly hydrophilic might show a particular higher position in the SDS-PAGE analysis when compared to a hydrophobic one. We did compare the migration patterns of SMOBIO’s Protein Markers with other brands, and we concluded that it was difficult to define “precision” due to the reasons mentioned above. Therefore, in the product description, we suggest our users to calibrate the MW against their interested proteins. Although it is impossible to define "precision" for molecular weight of proteins in SDS-PAGE, we did compare the migration pattern of pre-stained markers with unstained protein marker (Invitrogen MARK12) for calibration. It is concluded that the estimated molecular weight of SMOBIO’s pre-stained marker shows a curve matching well with that of unstained native proteins (MARK12), representing a good estimation of the MW of each pre-stained protein in the SDS-PAGE analysis.
Will SMOBIO’s Protein Markers/Ladder be washed out during Western blotting process?
SMOBIO’s Protein Markers/Ladder will be only slightly washed out during Western blotting process. However, the excess of Tween-20 (more than 0.2%) in washing buffer will affect SMOBIO’s Protein Markers/Ladder on the transfer membrane.
Here are suggestions for Western blotting process:
1. Transfer SMOBIO’s Protein Markers/Ladder to membrane with transfer buffer containing 20% methanol to fix SMOBIO’s Protein Markers/Ladder on membrane.
2. Wash membrane with PBS or TBS containing less than 0.1% Tween-20.
Will SMOBIO’s Protein Markers/Ladder be affected by the stripping/deprobing process with the presence of β-Mercaptoethanol (β-ME)?
In normal circumstances, the presence of βME during the stripping/deprobing process will only slightly affect SMOBIO’s Protein Markers/Ladder. However, the presence of Tween-20 on PVDF membrane during the stripping/deprobing process has adverse effects on SMOBIO’s Protein Markers/Ladder.
Here are suggestions for Western stripping/deprobing process:
1. Wash the PVDF membrane in methanol for 5~10 minutes prior to the stripping/deprobing process to mitigate the adverse effect of Tween-20.2. Recommended stripping buffer (for 1 L):
15 g glycine,
1 g SDS,
10 mL Tween 20.
Dissolve in 800 mL distilled water.
Adjust pH to 2.2
Bring volume up to 1 L with distilled water
Terence L Kirley, Andrew B Norman Biochem Biophys Rep. 2023 Jul 28;35:101520. doi:10.1016/j.bbrep.2023.101520. eCollection 2023 Sep.
PMCID: PMC10404603
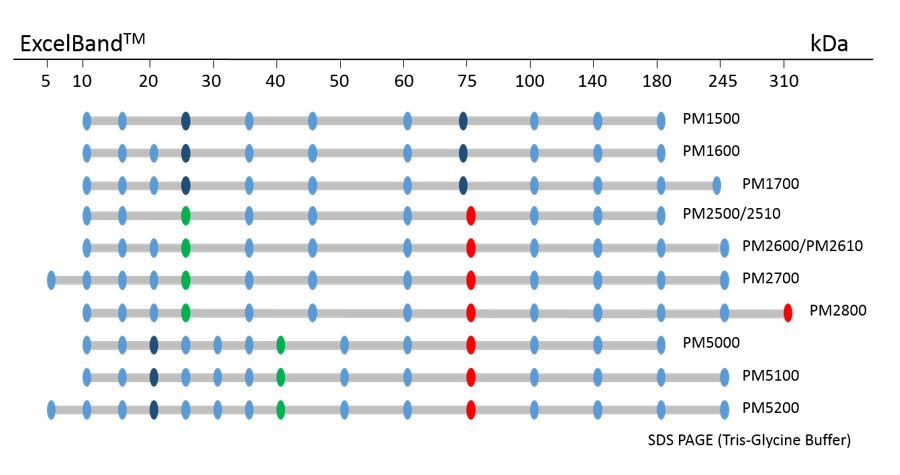
ExcelBand™ Protein Markers
Ready-to-use— premixed with a loading buffer for direct loading, no need to boil
Broad range— 310 kDa to 5 kDa
Pre-stained bands — for monitoring protein separation during electrophoresis and Western blotting transferring efficiency on membrane
Enhanced bands— for quick reference
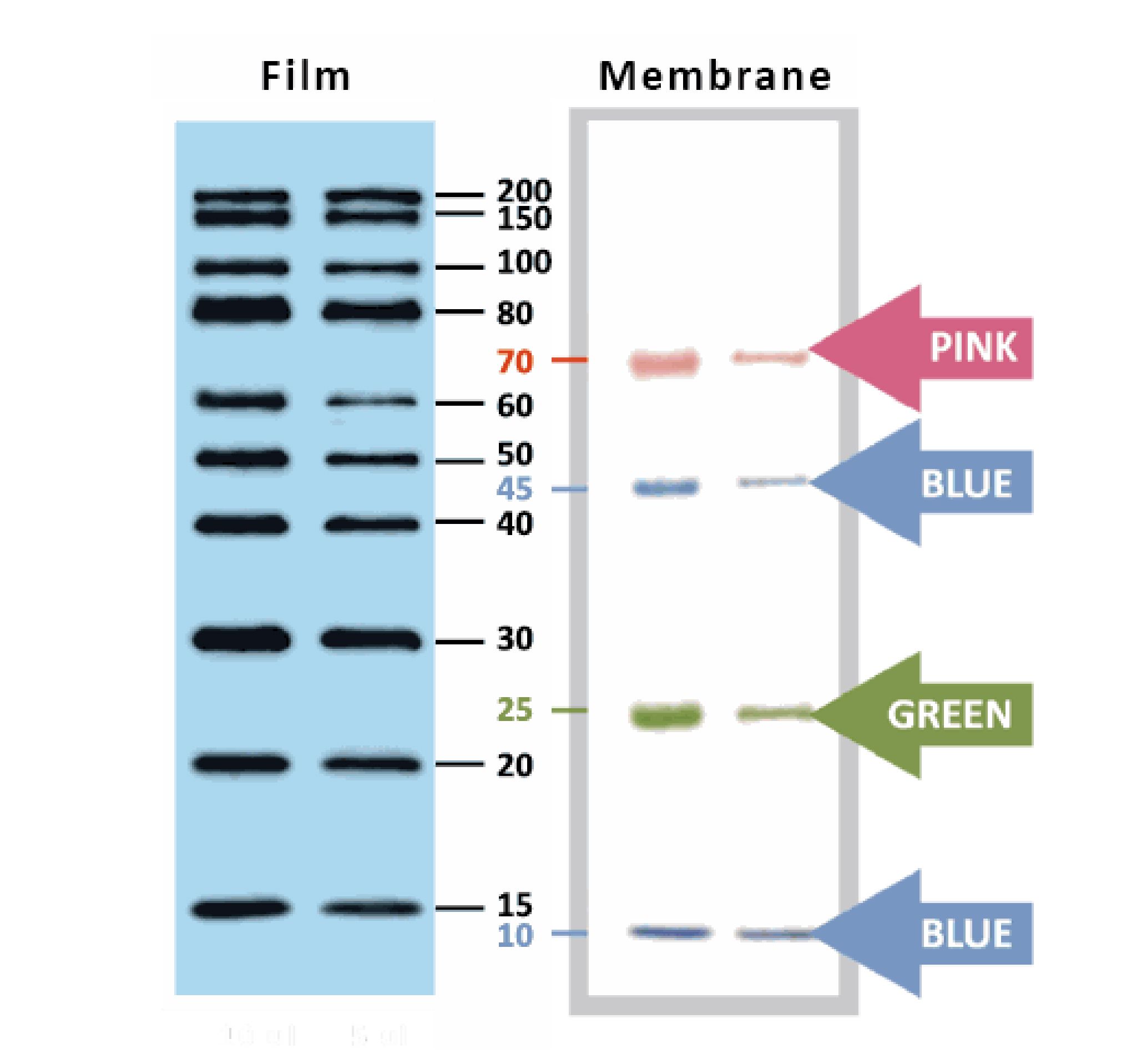
YesBlot™ Western Marker I
Ready-to-use — no need of mixing or heating before sample loading
Direct visualization — 10 IgG-binding proteins for direct visualization on Western blots
Pre-stained bands — 4 pre-stained proteins for monitoring protein separation during electrophoresis and Western blotting transferring efficiency on membrane
Wide range — 10 clear bands from 15 to 200 kDa for size estimation
Quick reference — two enhanced bands (30 and 80 kDa)
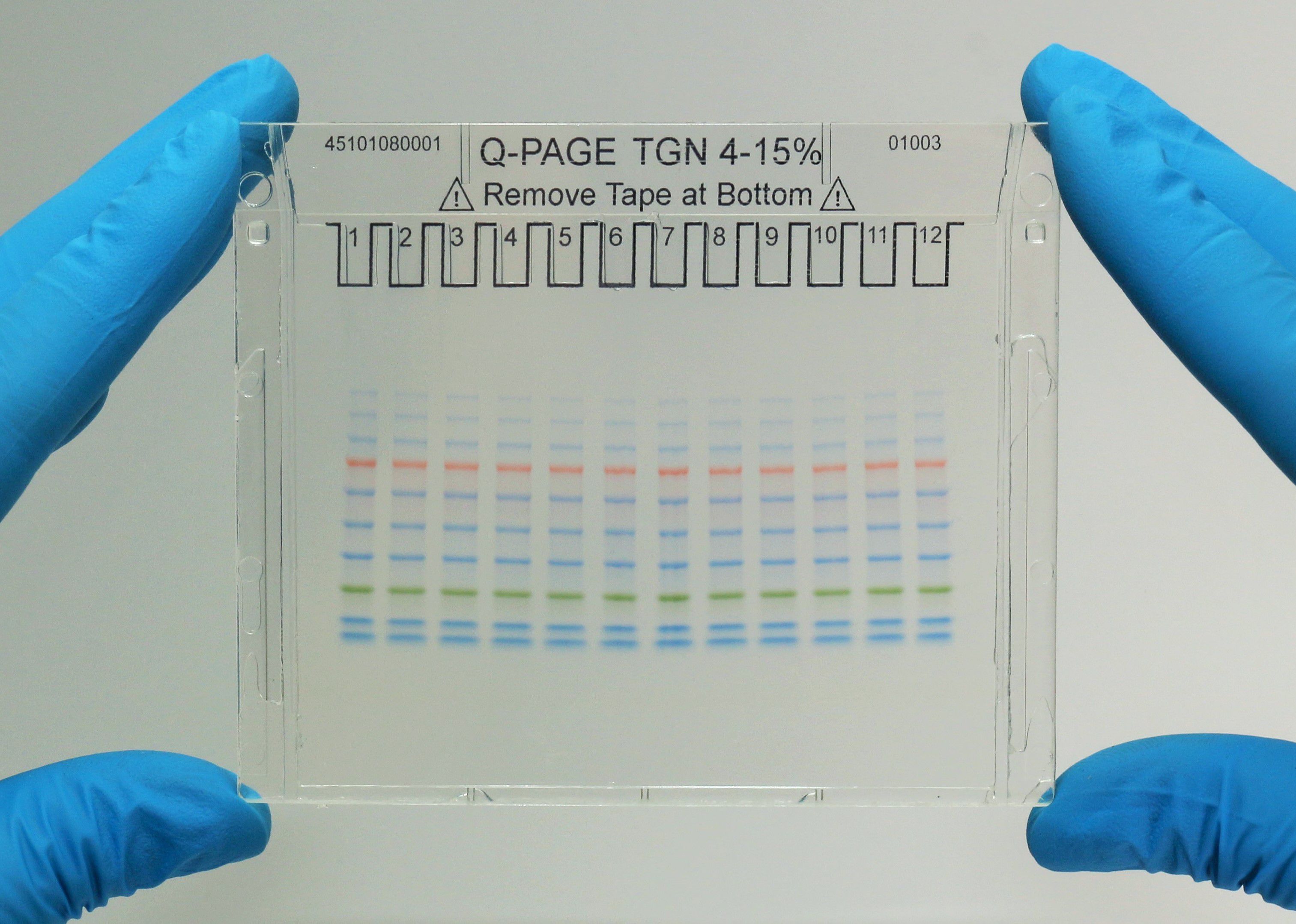
Q-PAGE™ Precast Gels
User-friendly gel cassette:
Numbered and framed wells for sample loading
Labeled warning sign and green tape as reminder
Enhanced gel performance:
Enhanced gel electrophoresis speed
Better band separation
Stable for shipping at ambient temperature
Easy compatibility:
Available as homogeneous and adjusted gradient gels for a wide range of protein separation.
Compatible with most popular protein electrophoresis systems

![[PM1600] ExcelBand™ All Blue Regular Range Plus Protein Marker (9-180 kDa), 250 μl x 2](/web/image/product.template/249/image?unique=5fb741a)
![[PM1600] ExcelBand™ All Blue Regular Range Plus Protein Marker (9-180 kDa), 250 μl x 2](/website/image/product.template/249/image/90x90)